Bistability is a fundamental property of engineering and natural systems, with the ability to
switch and maintain states. Prokaryotes couple the input and utilization of metabolites with
feedback-regulated genetic circuits by constructing bistable switches,
using inducible transcriptional and enzymatic components to create hybrid systems, which allow
us to regulate the pathway easily[1]. All bistable switches developed so far, however, control
the expression of target genes without
access to other layers of the cellular machinery[2]. The binding of repressor proteins and
operons can lead to incomplete pathway repression due to different selection and concentration
of inducers, which affects the regulation
of metabolic pathways by bistable switches[3]. Our experiment enhances the binding strength
between the repressor protein and DNA by optimizing the switch[4], achieving lower basal level
expression, induced high-level expression,
and ultimately achieving bistable expression of the target gene.
This method has great potential to expand the functionality of biomolecular devices and can be
used to separate growth and production processes during fermentation. Based on the specificity
of hyaluronic acid produced by multiple metabolic pathways, we
applied the designed bistable switch to regulate the carbon flow direction of engineering
bacteria fermenting hyaluronic acid and tested the degree of performance improvement after
optimizing the switch. We attempted to construct
a bistable switch using ordinary Escherichia coli. Due to the production of hyaluronic acid by
the precursors UDP-glucuronic acid and UDP N-acetyl-glucosamine in the glucose metabolism
pathway of E. coli under the action of hasA,
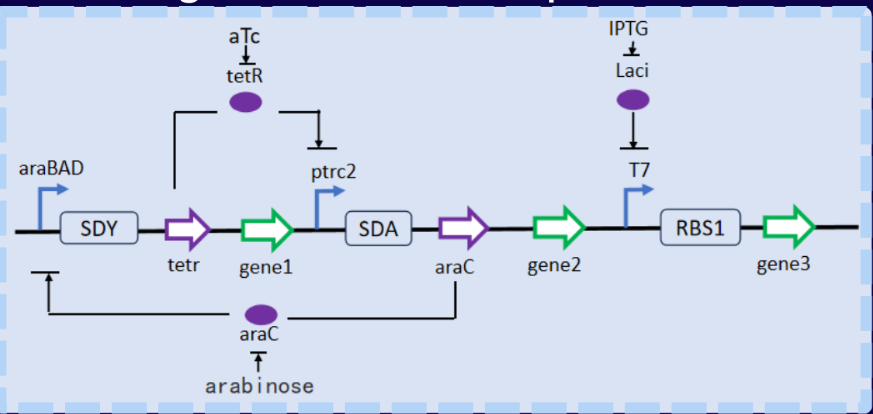
we plan to use arabinose operons to regulate the galU and udg gene pathways in the
UDP-glucuronic acid biosynthesis pathway of E. coli; In the biosynthesis pathway of
UDP-N-acetyl-glucosamine, tetracycline operons are used to regulate
gene pathways such as glmU, glmS, and glmM[5]. We plan to regulate the ratio of opening of the
two pathways by changing the concentration of added inducers arabinose and dehydrated
tetracycline, and during the growth process, make
the metabolic pathway more inclined towards growth-related pathways, allowing the strain to grow
rapidly first. During the fermentation process, the metabolic pathway is biased towards the
pathway that produces fermentation products,
allowing the strain to produce a large amount of fermentation products. In the testing phase, we
used GFP instead of the relevant genes in order to more accurately and quickly detect the
performance of the mutants. After the design
of the bistable switch was completed, we used error-prone PCR and computer simulation to evolve
araC and tetR genes directly, to optimize the bistable switch, increase its ability to inhibit
protein binding to DNA, reduce promoter
leakage, and increase the performance of the bistable switch.
[1] Huang D, Holtz WJ, Maharbiz MM. A genetic bistable switch utilizing nonlinear protein
degradation. J Biol Eng. 2012 Jul 9;6(1):9. https://doi.org/10.1186/1754-1611-6-9 PMID:
22776405; PMCID: PMC3439342.
[2] Oyarzún DA, Chaves M. Design of a bistable switch to control cellular uptake. J R Soc
Interface. 2015 Dec 6;12(113):20150618. https://doi.org/10.1098/rsif.2015.0618 PMID: 26674196;
PMCID: PMC4707844.
[3] Rolf Lutz, Hermann Bujard, Independent and Tight Regulation of Transcriptional Units in
Escherichia Coli Via the LacR/O, the TetR/O and AraC/I1-I2 Regulatory Elements, Nucleic Acids
Research, Volume 25, Issue 6, 1 March 1997, Pages 1203–1210,
https://doi.org/10.1093/nar/25.6.1203
[4] von Hippel PH, Revzin A, Gross CA, Wang AC. Non-specific DNA binding of genome regulating
proteins as a biological control mechanism: I. The lac operon: equilibrium aspects. Proc Natl
Acad Sci U S A. 1974 Dec;71(12):4808-12. https://doi.org/10.1073/pnas.71.12.4808
PMID: 4612528; PMCID: PMC433986.
[5] Woo JE, Seong HJ, Lee SY, Jang YS. Metabolic Engineering of Escherichia coli for the
Production of Hyaluronic Acid From Glucose and Galactose. Front Bioeng Biotechnol. 2019 Nov
21;7:351. https://doi.org/10.3389/fbioe.2019.00351 PMID: 31824939; PMCID:
PMC6881274.
