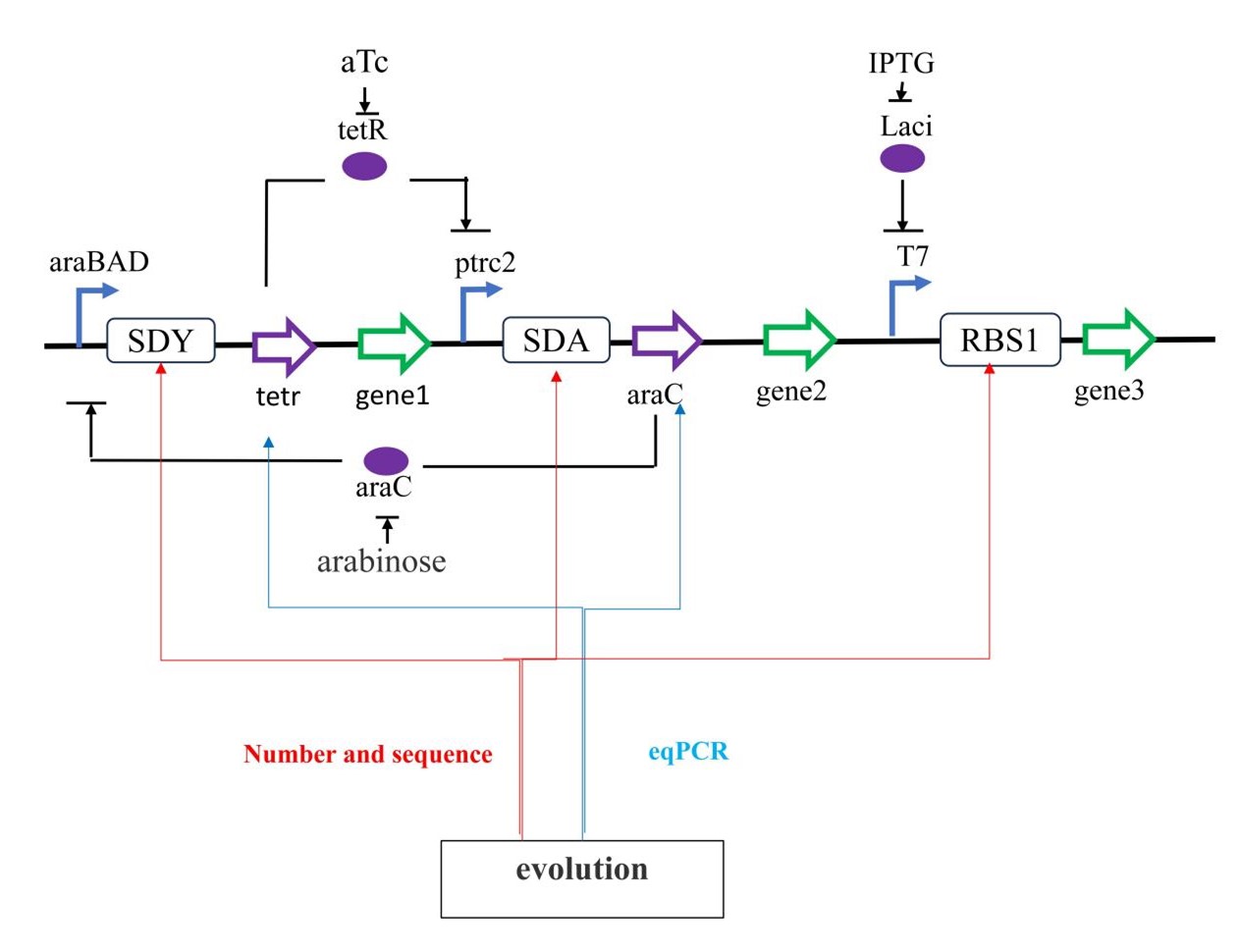
We have built a bistable switch to control the rate at which cells take up a metabolite from the
environment[1] and are preparing to mutate the araC and tetR genes responsible for regulating
the switch. Our goal is to screen for the most effective and
sensitive switches by detecting downstream toxic proteins and fluorescent signals to aid in
different applications of bistable switches(figure 12).
Gene regulatory switch. Act as a sign of evolution gene3 can be replaced with GFP . The change
of gene1, gene2 and gene3 can be used to regulate various metabolic production pathways.Directed
evolution of the ribosome binding site may also be one of the
ways to optimize the switch
Other Researchers have knocked out genes such as pfkA, pfkB, zwf, galR, and galS in E. coli, and
introduced galU and kfiD genes to enable them to activate the galactose pathway and reduce the
amount of glucose entering respiratory metabolism, enabling
them to efficiently utilize glucose and galactose to synthesize the rate limiting product
UDP-glucuronic acid in the hyaluronic acid production pathway, thereby efficiently synthesizing
hyaluronic acid. [2] Our designed switch
aims to further optimize this strategy. We will conduct experiments in E. coli producing HA to
achieve artificial control between the cell's growth and production modes.
We have spent a lot of time modifying E. coli k12 to enable it to undergo the next step of
directed evolution. However, we did not successfully transfer the designed plasmid into E. coli
K 12. Many times of DNA agarose gel recovery, digestion and reconnection
have made the concentration of our target DNA segments too low. However, after computer
prediction, we believe that the introduction of gene regulation switches into the directed
evolution process may help the biosynthesis of E.
coli.
The algorithm we constructed for predicting whether a protein is a DNA binding protein and
evaluating its binding strength, and used it to simulate mutations in AraC and TetR proteins to
improve protein DNA binding efficiency, reduce switch leakage, and
increase switch performance.
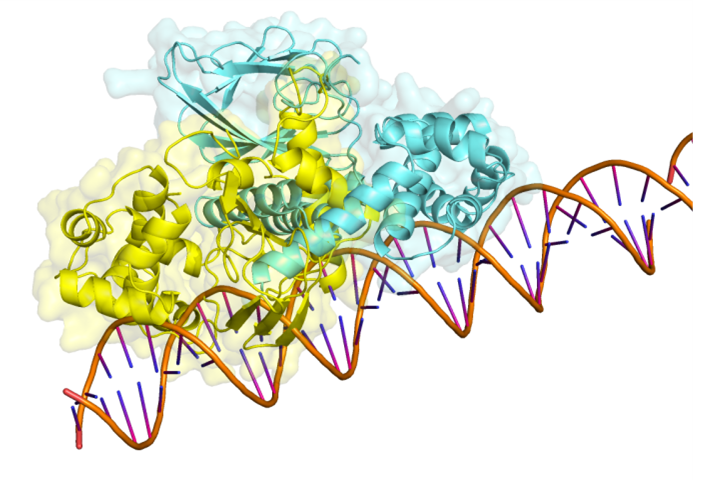
AraC-DNA binding site before (yellow) and after (blue) mutation.
Since the algorithm cannot specify the site where the protein binds DNA, certain changes to the
DNA binding site occur before and after AraC mutation (Figure 8), so the algorithm needs to
control the number of mutation sites to reduce the influence of
binding site movement. Although the AraC and DNA binding sites are changed, they are still in
the region of the operon-binding repressor protein, so the model can be considered valid
[1] Oyarzún DA, Chaves M. Design of a bistable switch to control cellular uptake. J R Soc
Interface. 2015 Dec 6;12(113):20150618. https://doi.org/10.1098/rsif.2015.0618 PMID: 26674196;
PMCID: PMC4707844.
[2] Woo JE, Seong HJ, Lee SY, Jang YS. Metabolic Engineering of Escherichia coli for the
Production of Hyaluronic Acid From Glucose and Galactose. Front Bioeng Biotechnol. 2019 Nov
21;7:351. https://doi.org/10.3389/fbioe.2019.00351 PMID: 31824939; PMCID:
PMC6881274.