Method Dry Lab
Rational design of the mutants
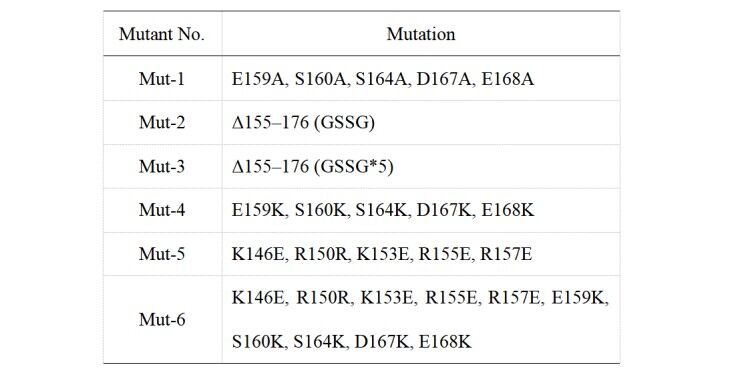
Table 1. Mutation sites of different mutants
Molecular dynamics simulation
The simulation time was 400 ns and result is calculated based on a simulated track by using the program provided by GROMAC 5.2.11 software. The GROMOS96 force field is selected, and the SPC model is adopted for water molecules. The Lenard Jones function is used to calculate the van der Waals force. The non-bond truncation distance is set to 0.9 nm. The list of non-bond atoms is updated every four steps. The particle-mesh Ewald method is used to calculate the electrostatic interaction. The Verlet leapfrog algorithm is used to solve the equation of motion at each step, and the coordinates of each atom at the new time are obtained through integration, which is 2 fs. At the same time, the LINCS algorithm is used to fix the relative distance between all bonding atoms to reduce the computational complexity. All simulations were carried out in an isothermal isobaric ensemble (NPT) at a temperature of 300 K and 1.01 × 105 Pa. Berendsen method is adopted for temperature and pressure control, the time constant was set to 0.1 ps for temperature control and the pressure coupling constant is 0.5 ps, the continuous operation is 400 ns. All molecular dynamics simulation calculations are completed on our laboratory server. According to the gmx rms module in the Gromacs package, we calculate the RMSD of different components in the simulation system. We analyzed the changes of six mutants in the system, and analyze the changes in the skeleton carbon atom of the protein and the central structure of the entire protein.