Description
Compared with Cas12a, CasΦ shows better mismatch target recognition capability. When a single-base mismatch is located in the center of the target recognition region, CasΦ nuclease shows a good substrate specificity for matched and mismatched targets. However, when the mismatched base moved to both sides, the non-specific cleavage increased significantly. This may be related to the effect of mismatched base position on the stability of RNA and mismatched target complex.
The results from Jennifer Doudna’s group showed that the helix α7 in CasΦ protein has a significant effect on enzyme activity. Moreover, recent studies have shown that the introduction of a hairpin secondary structure at the end of the spacer region of crRNA can reduce the affinity of Cas nuclease to crRNA/mismatched target complex, thereby improving the target recognition specificity of the CRISPR-Cas system. Our team hoped to further explore the mutations in helix α7 of CasΦ nuclease and combine them with hairpin structured crRNA to establish a both sensitivity and specificity improved CRISPR-Cas system for nucleic acid detection.
Based on the above, we modified the protein by point mutation. Using GROMAC 5.2.11 software, LINCS algorithm, and other methods to carry out molecular dynamics simulation for the mutants, our team got four point mutations. With two mutants designed by Jennifer Doudna’s group, we expressed six mutants and verified their activity, with the hope to acquire mutants with enhanced even excellent performance.
The results from Jennifer Doudna’s group showed that the helix α7 in CasΦ protein has a significant effect on enzyme activity. Moreover, recent studies have shown that the introduction of a hairpin secondary structure at the end of the spacer region of crRNA can reduce the affinity of Cas nuclease to crRNA/mismatched target complex, thereby improving the target recognition specificity of the CRISPR-Cas system. Our team hoped to further explore the mutations in helix α7 of CasΦ nuclease and combine them with hairpin structured crRNA to establish a both sensitivity and specificity improved CRISPR-Cas system for nucleic acid detection.
Based on the above, we modified the protein by point mutation. Using GROMAC 5.2.11 software, LINCS algorithm, and other methods to carry out molecular dynamics simulation for the mutants, our team got four point mutations. With two mutants designed by Jennifer Doudna’s group, we expressed six mutants and verified their activity, with the hope to acquire mutants with enhanced even excellent performance.

Figure. 1 Maximum likelihood phylogenetic tree of type V subtypes a-k. Phage-encoded CasΦ proteins are outlined in red, with prokaryote and transposon-encoded proteins in blue. Bootstrap and approximate likelihood ratio test values >90 are shown on the branches (circles)[1].
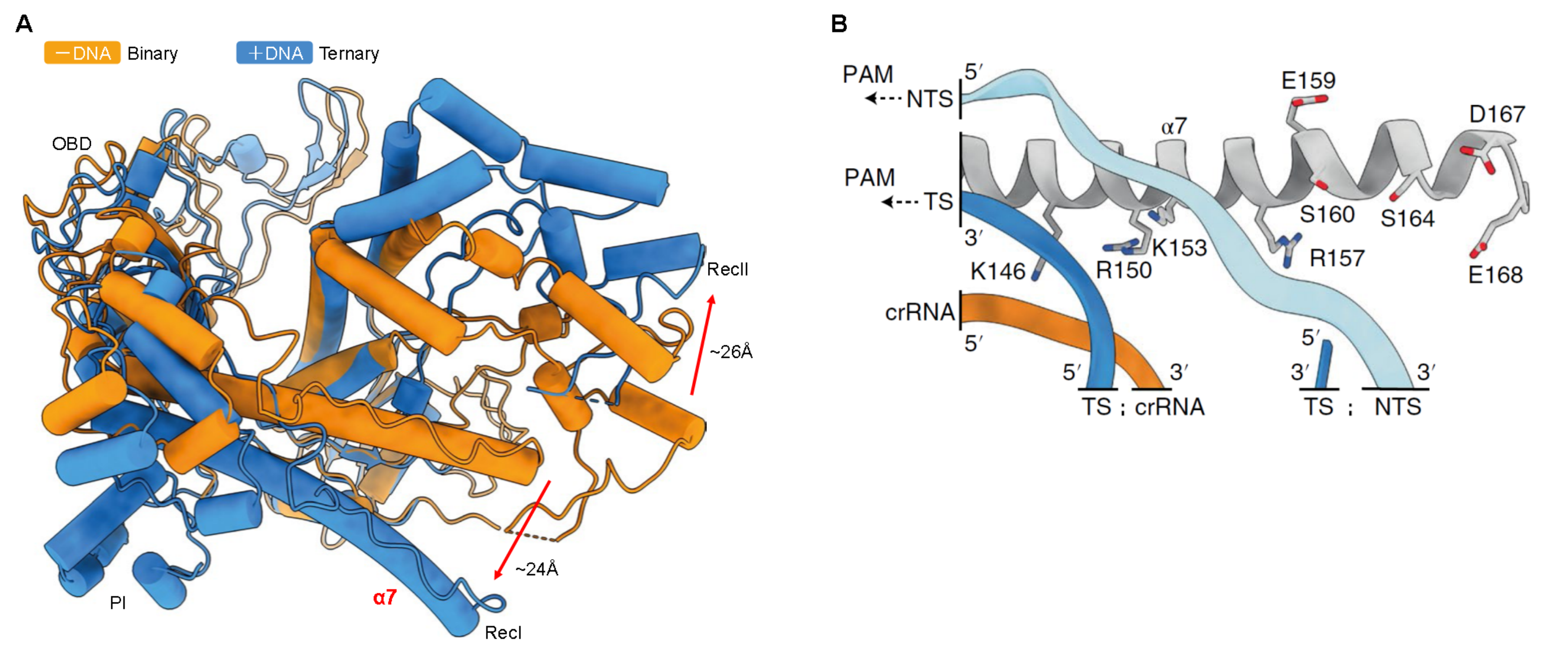
Figure. 2 Structural diagram of CasΦ protein[2]. A: Superimposition of the binary state (orange) and ternary state (blue) highlighting the rearrangement (arrows) of RecI and RecII. OBD:
Oligonucleotide-binding domain; PI: PAM-interacting domain; Rec: recognition structure. B: Detailed view of the nucleic acids and helix α7. Positively charged amino acids are marked in blue and negatively charged amino acids are marked in red.
Reference:
[1]Sapranauskas, R., G. Gasiunas, C. Fremaux, etal., The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res, 2011. 39(21): p. 9275-82.doi: 10.1093/nar/gkr606.
[2]Pausch, P., K.M. Soczek, D.A. Herbst, et al., DNA interference states of the hypercompact CRISPR-CasPhi effector. Nat Struct Mol Biol, 2021. 28(8): p. 652-661.doi: 10.1038/s41594-021-00632-3.
[2]Pausch, P., K.M. Soczek, D.A. Herbst, et al., DNA interference states of the hypercompact CRISPR-CasPhi effector. Nat Struct Mol Biol, 2021. 28(8): p. 652-661.doi: 10.1038/s41594-021-00632-3.

