

FIGURE
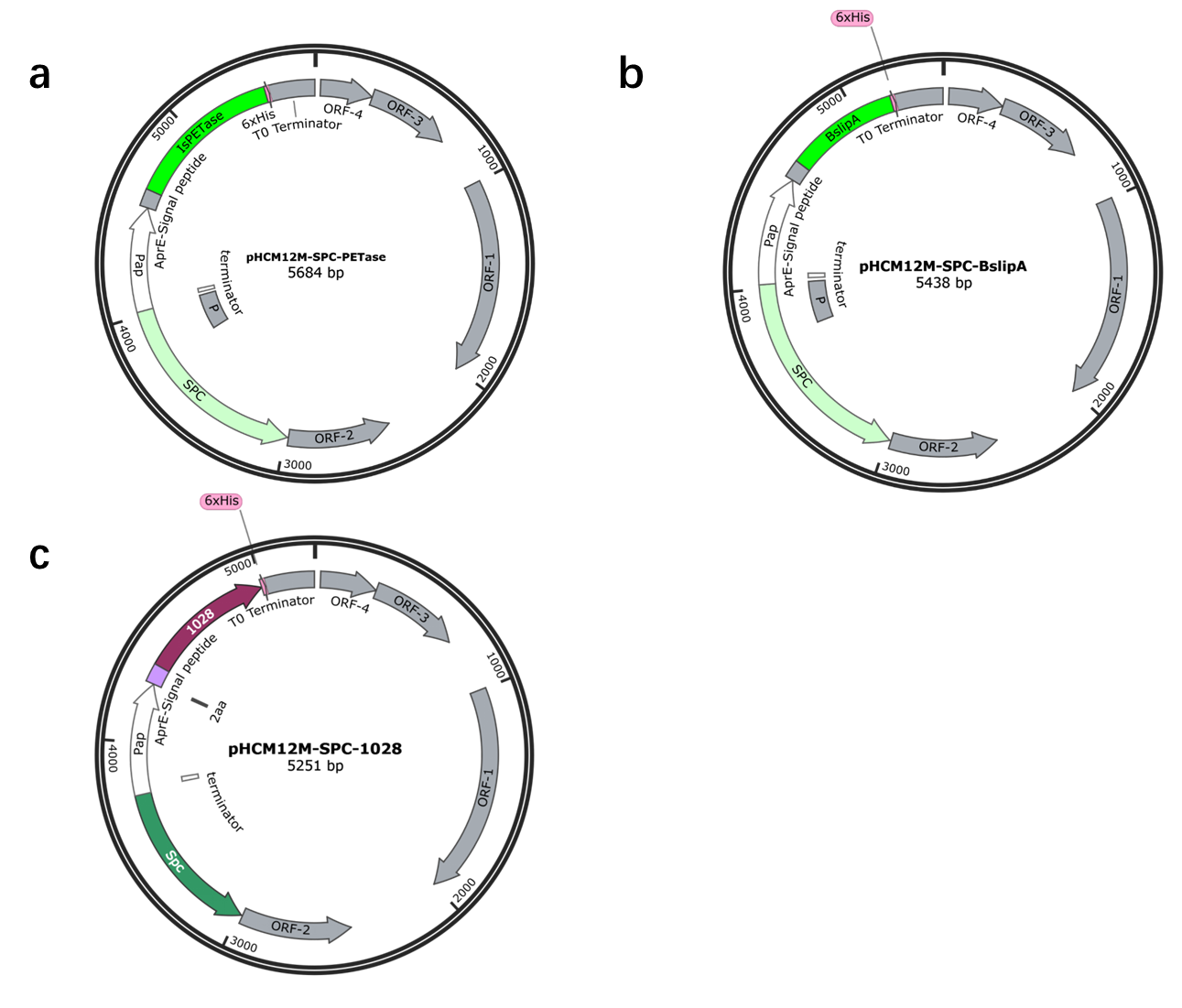
Figure S1. The map of plasmids used for directed evolution of recombinant plasmids. (A) pHCM12M-SPC-PETase . This plasmid contains the gene of IsPETase. (B) pHCM12M-SPC-BsLipA. The plasmid contains the gene of BsLipA. (C) pHCM12M-SPC-1028. The plasmid contains the gene of Lipase1028.
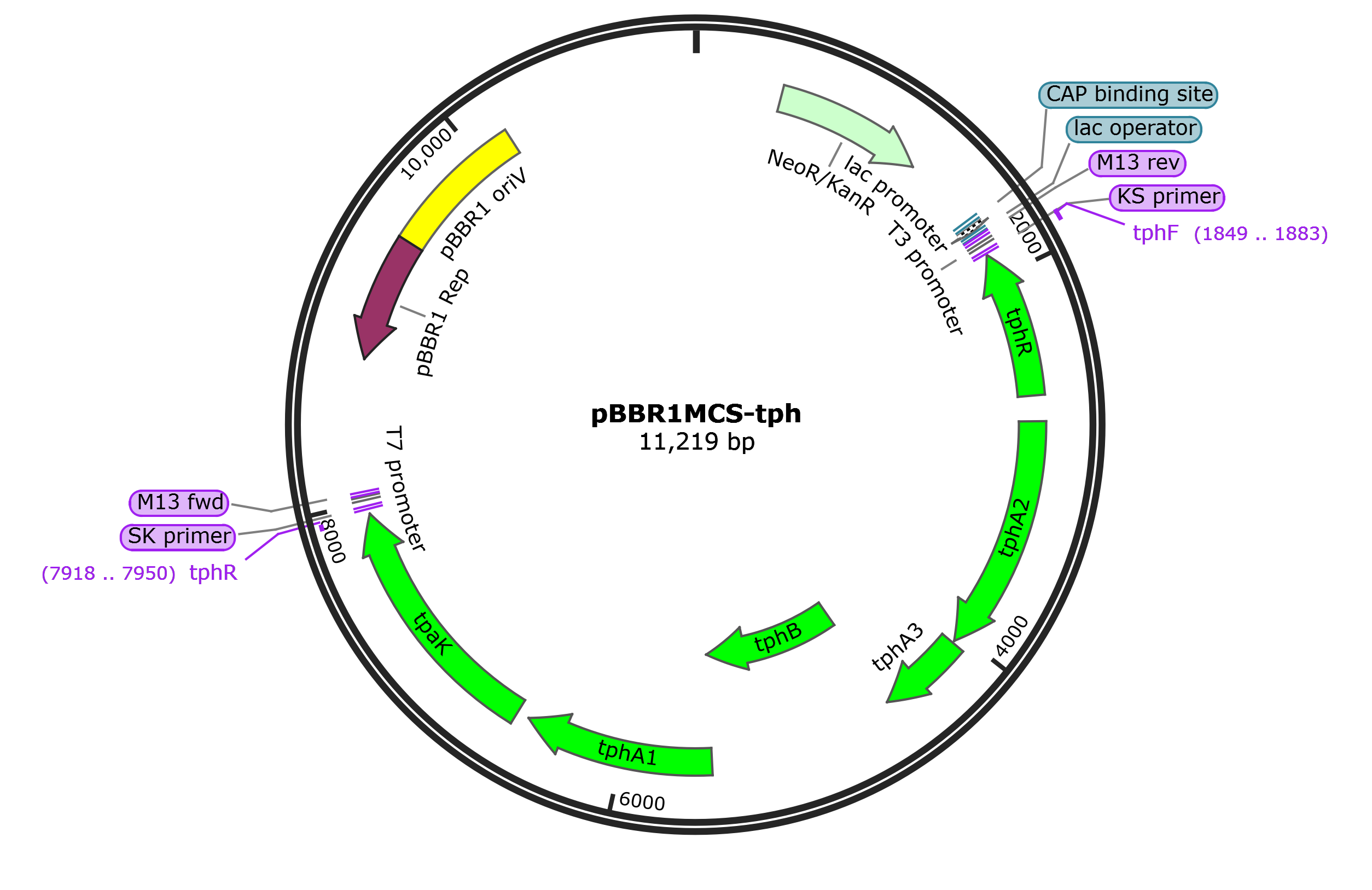
Figure S2. The map of pBBR1MCS-tph plasmids for KT2440 strain modification. This plasmid contains the tph clusters including genes encoding the transcriptional regulator (TphR), TPA transporter (TpaK), TPA 1, 2-dioxygenase (TphA), and 1, 2-dihydroxy-3, 5-cyclohexadiene-1, 4-dicarboxylate dehydrogenase (TphB) from a TPA degrading P. Stutzeri.
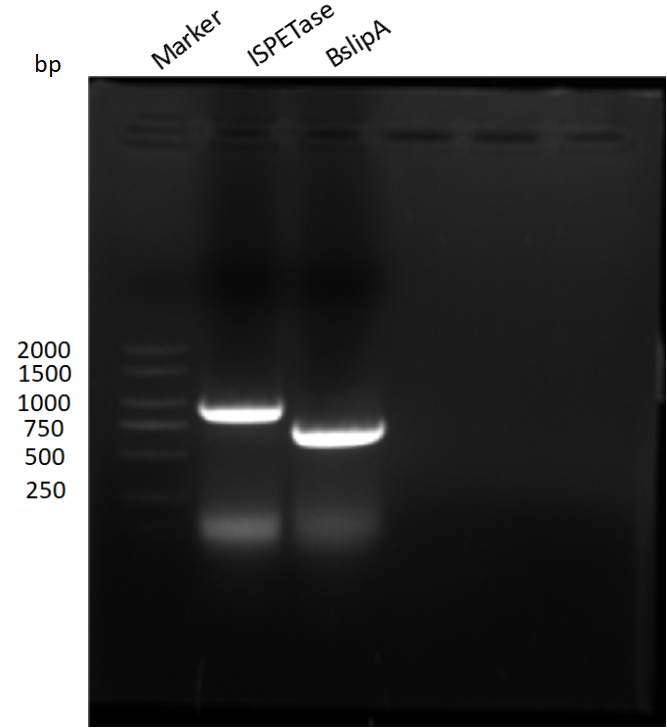
Figure S3. The DNA bands of IsPETase and BsLipA performing agarose gel (1 %) electrophoresis (120V, 30min). The correct band length for IsPETase is 789 bp and for BsLipA is 543 bp.

Figure S4. LB Petri dishes for constructing IsPETase and BsLipA libraries. Morphological changes of PBAT films after degradation.

Figure S5. PCR products generated by the overlap extension PCR. The highlights in the gel tray is the POE-PCR product.
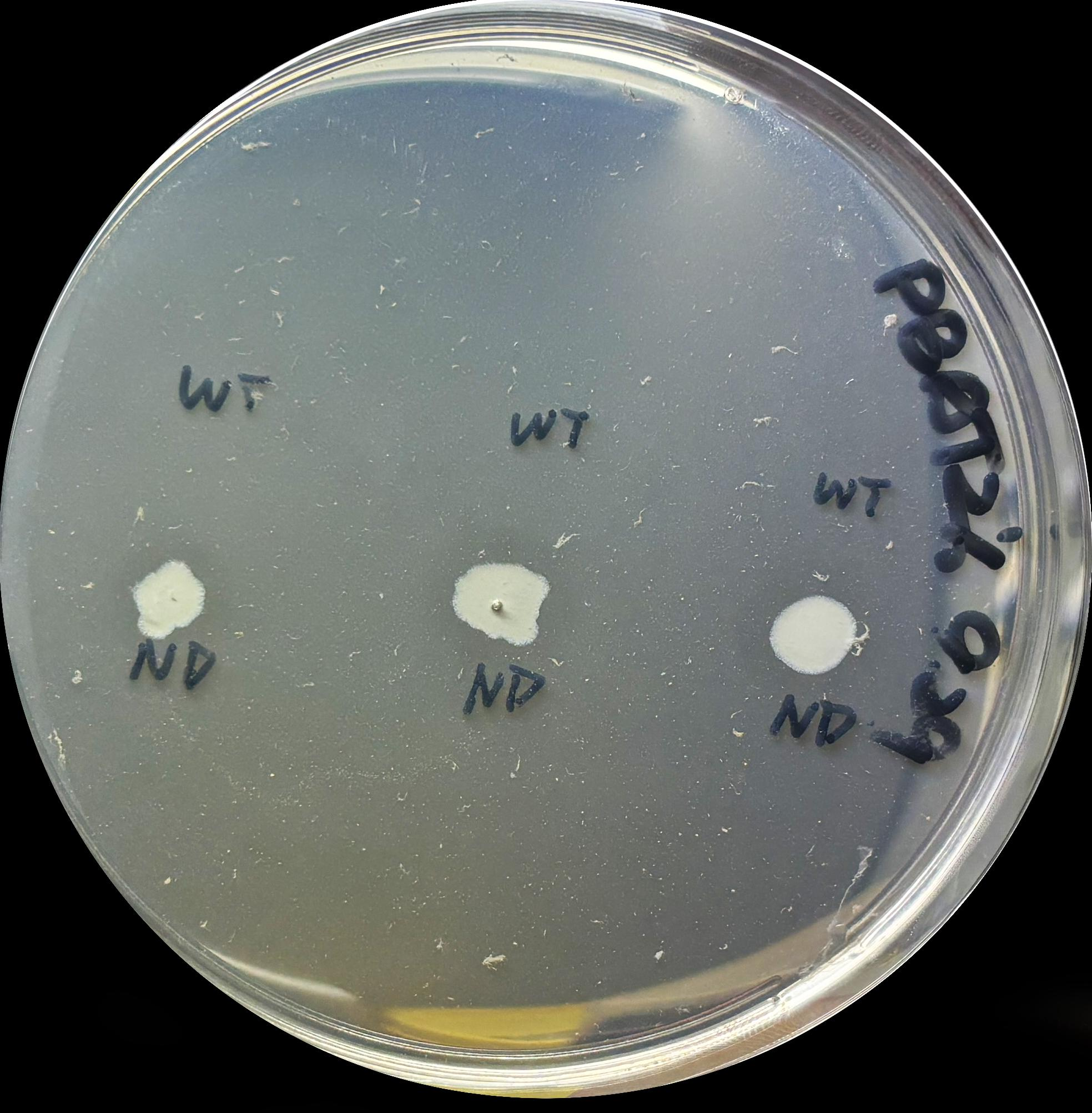
Figure S6. Hydrolysis zones of ND within 24h.
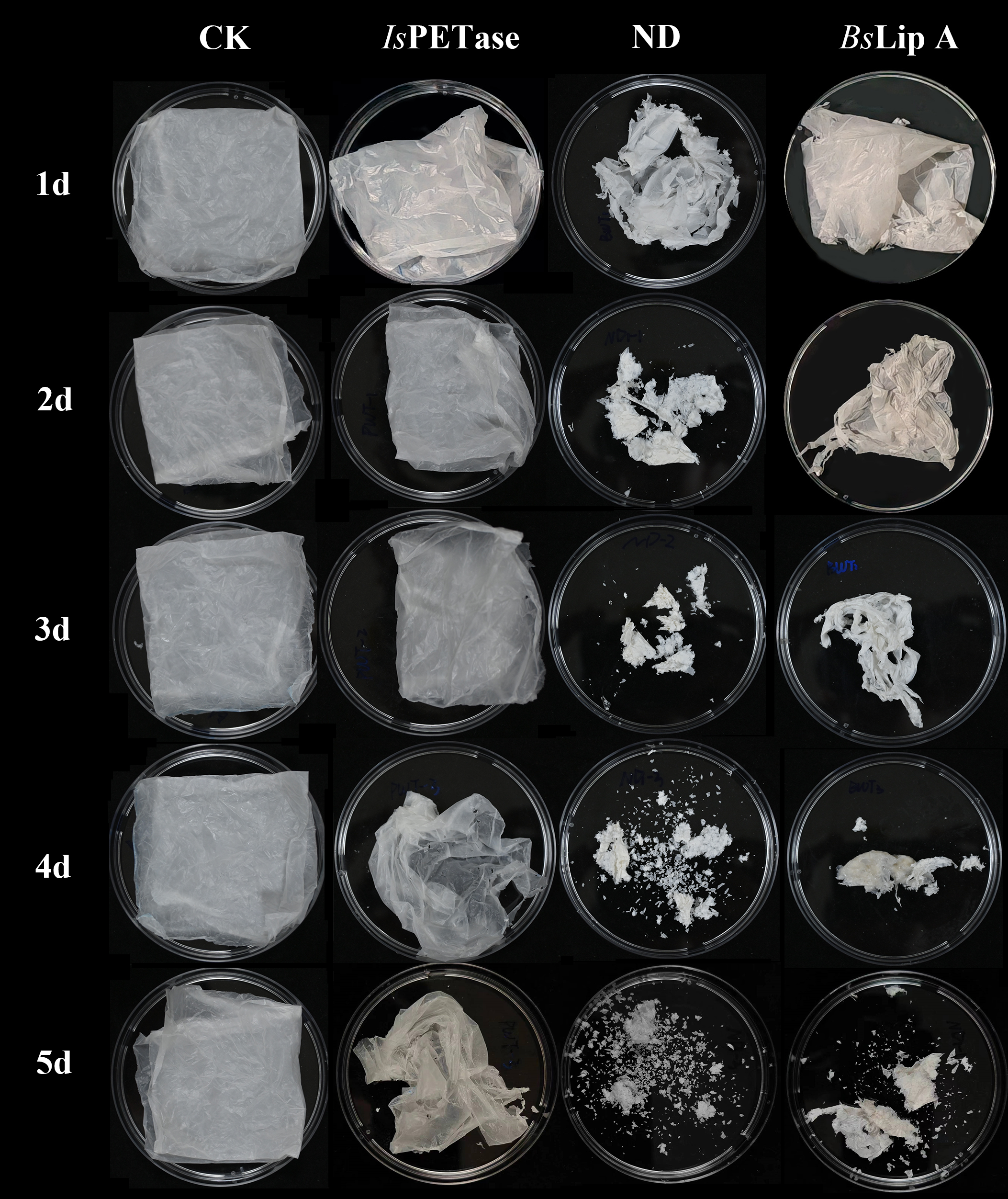
Figure S7. Morphological changes of PBAT films over different degradation periods. After incubating for 5 days in LB liquid medium without inoculated bacteria, the control group's PBAT remained intact, showing no significant changes. In the treatment group, PBAT films exhibited increasing degradation rates, turning whiter in color and decreasing in ductility when co-incubated with different strains. Films treated with IsPETase showed relatively minor changes, with a slight reduction in overall film area and a few holes appearing on the surface. However, films treated with BsLipA and ND strains underwent significant degradation over time, cracking into pieces, reducing in area, and eventually degrading into almost powder form. Notably, the films treated with ND strain had the highest degradation rate.
Figure S8. The three-dimensional structure of BABTaB.
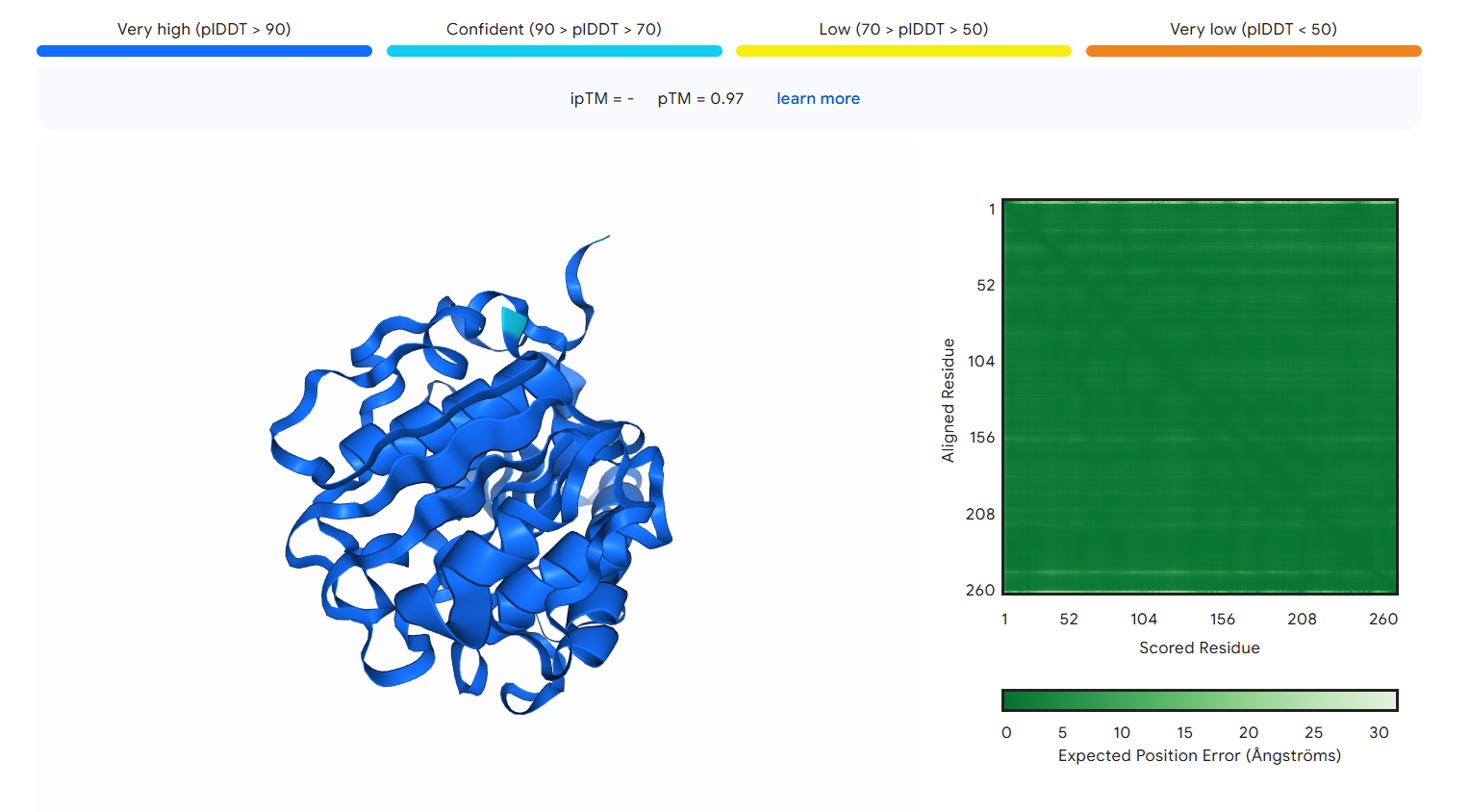
Figure S9. The three-dimensional structure of IsPETase.
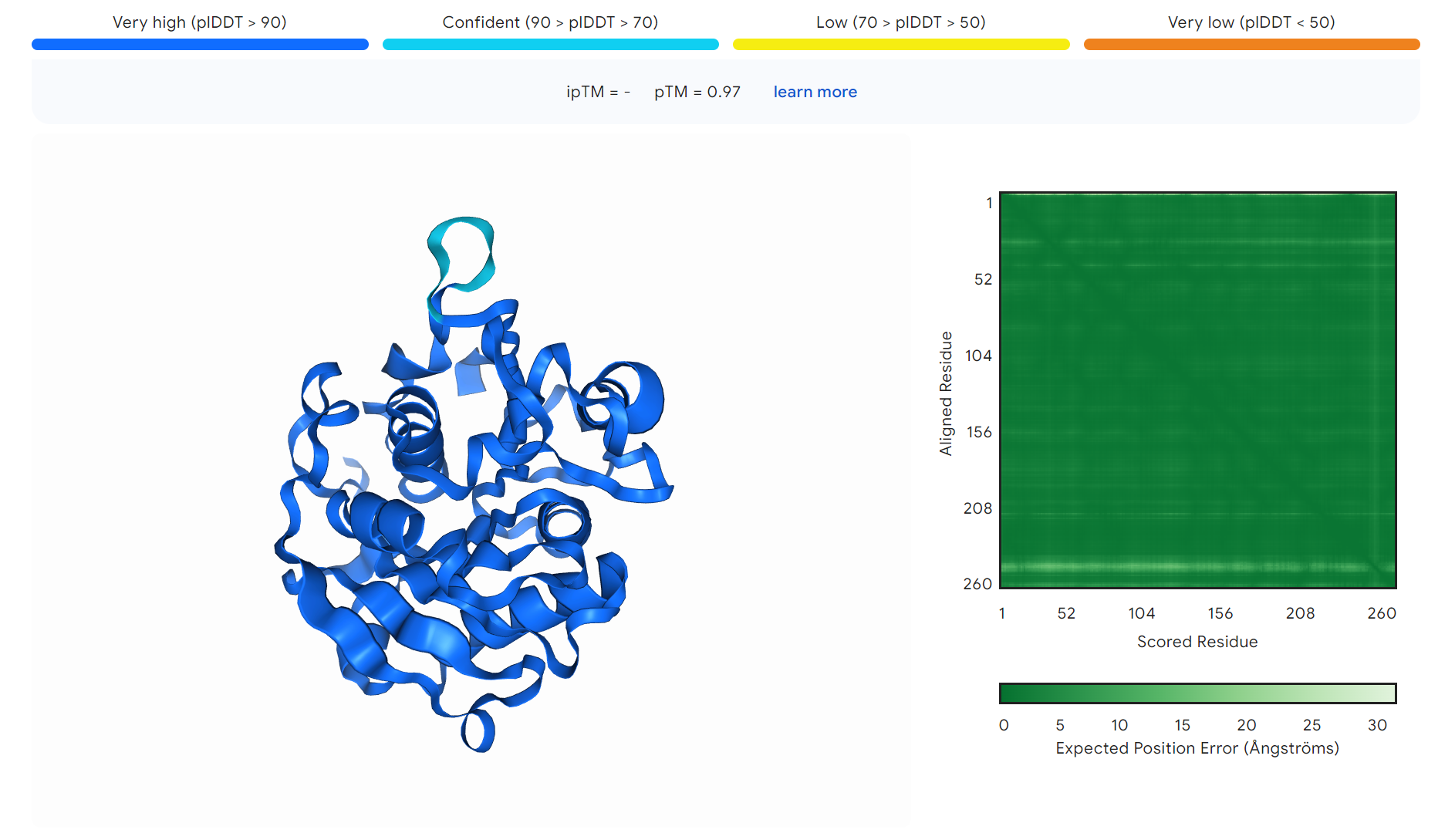
Figure S10. The three-dimensional structure of ND.
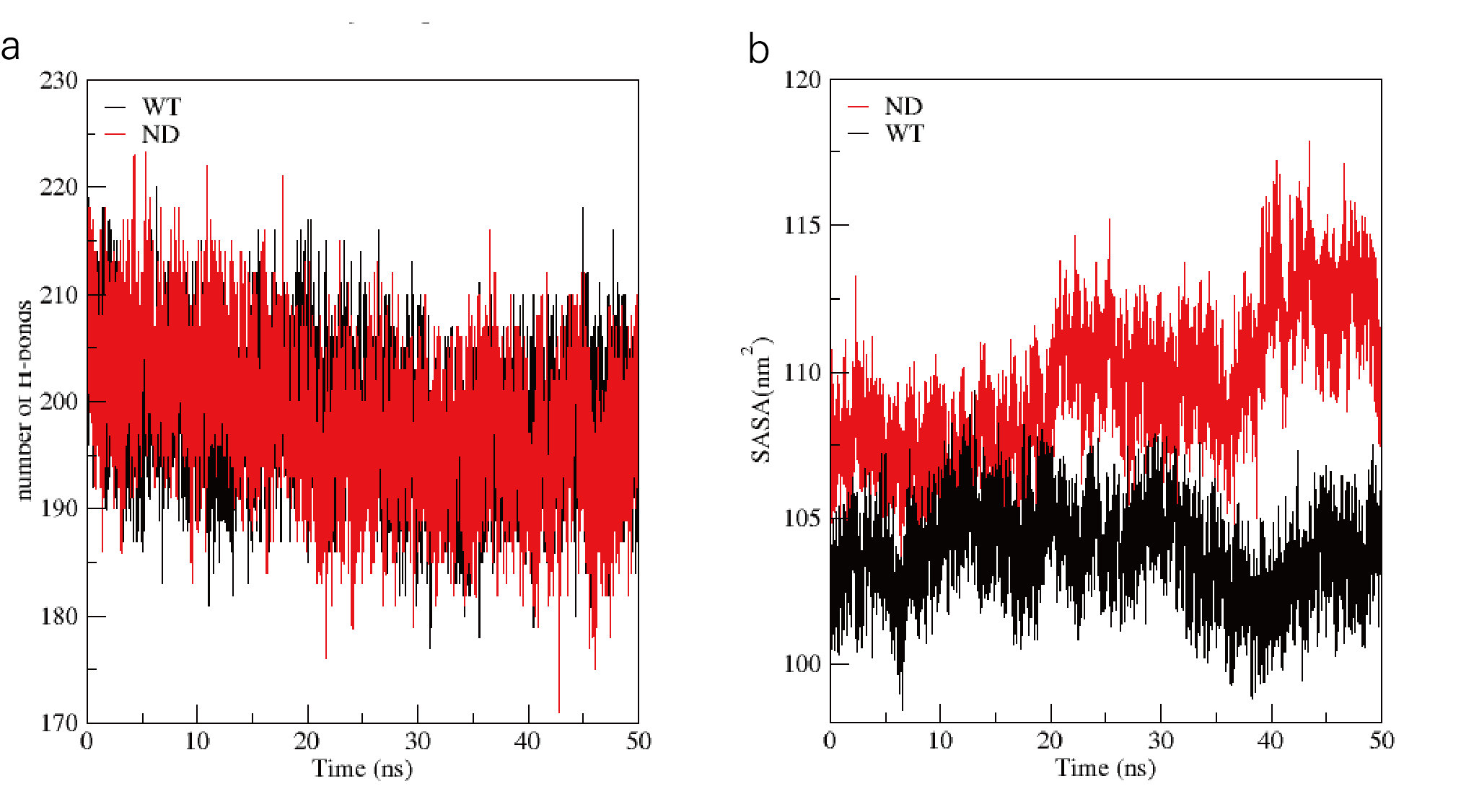
Figure S11. Number of H-bonds and SASA of IsPETase and mutant ND. (a) The number of hydrogen bonds during the protein-ligand complex simulation process;(b) The SASA curve of the protein during the protein-ligand complex simulation process (The solvent accessible surface area (SASA) is calculated by the van der Waals interaction between the solute and the solvent molecules. The SASA of the protein decreases as the protein density increases, so the change of SASA can predict the change of protein structure).
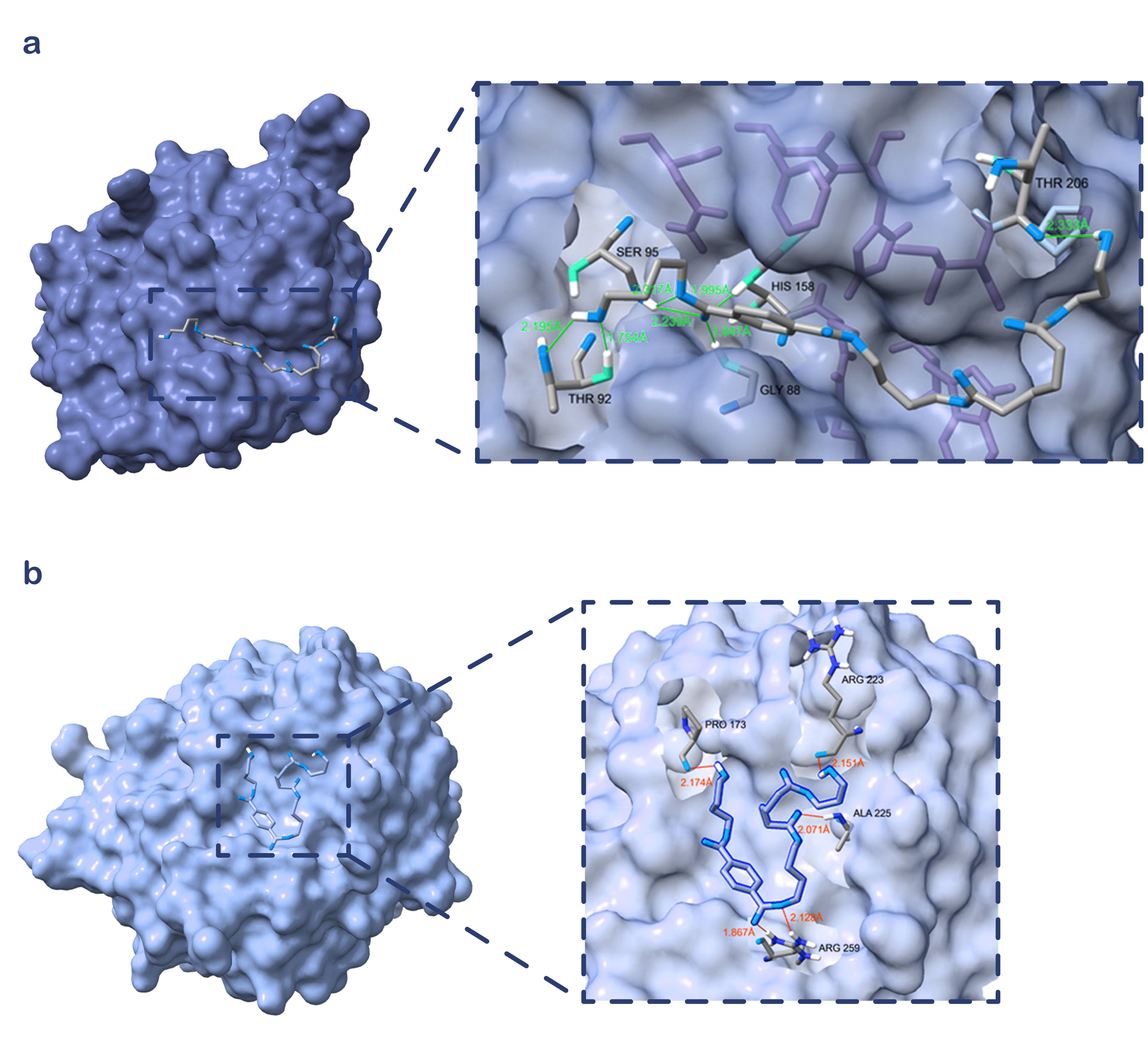
Figure S12. Molecular docking analysis of ND and IsPETase. (a) Hydrogen bond interactions between the mutant ND and BABTaB. The green dashed lines represent hydrogen bonds, with the numbers above representing the lengths of the hydrogen bonds; (b) Hydrogen bond interactions between the IsPETase and BABTaB. The amino acid residues involved in hydrogen bond interactions within the proteins are labeled and displayed in a stick model. The orange dashed lines indicate hydrogen bonds, with the numbers above representing the lengths of the hydrogen bonds.
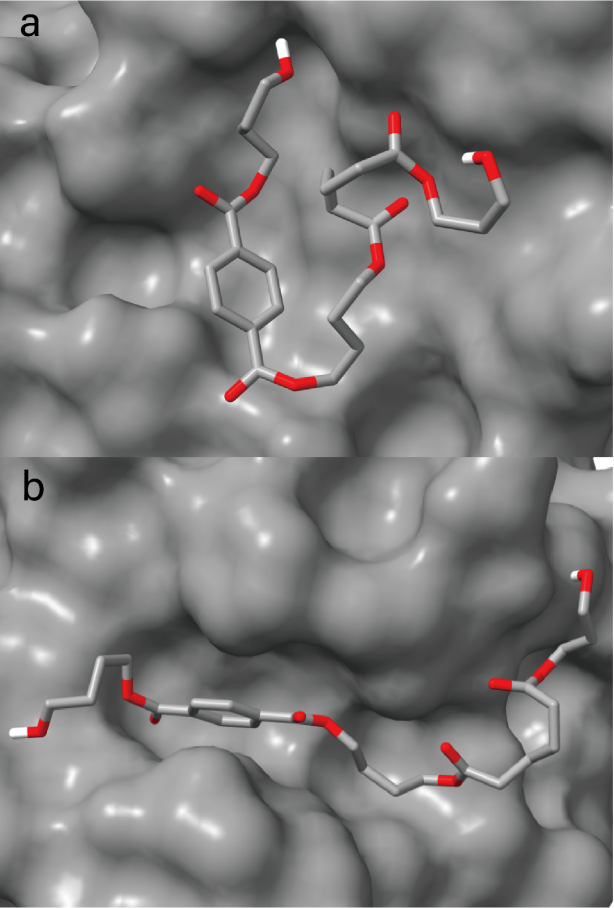
Figure S13. (a) The superimposed active site clefts of
BABTaB-bound complex of IsPETase; (b) The superimposed active site clefts
of BABTaB-bound complex of ND; The
BABTaB-bound pocket is shown in a surface model and the BABTaB molecule is
shown as a stick.
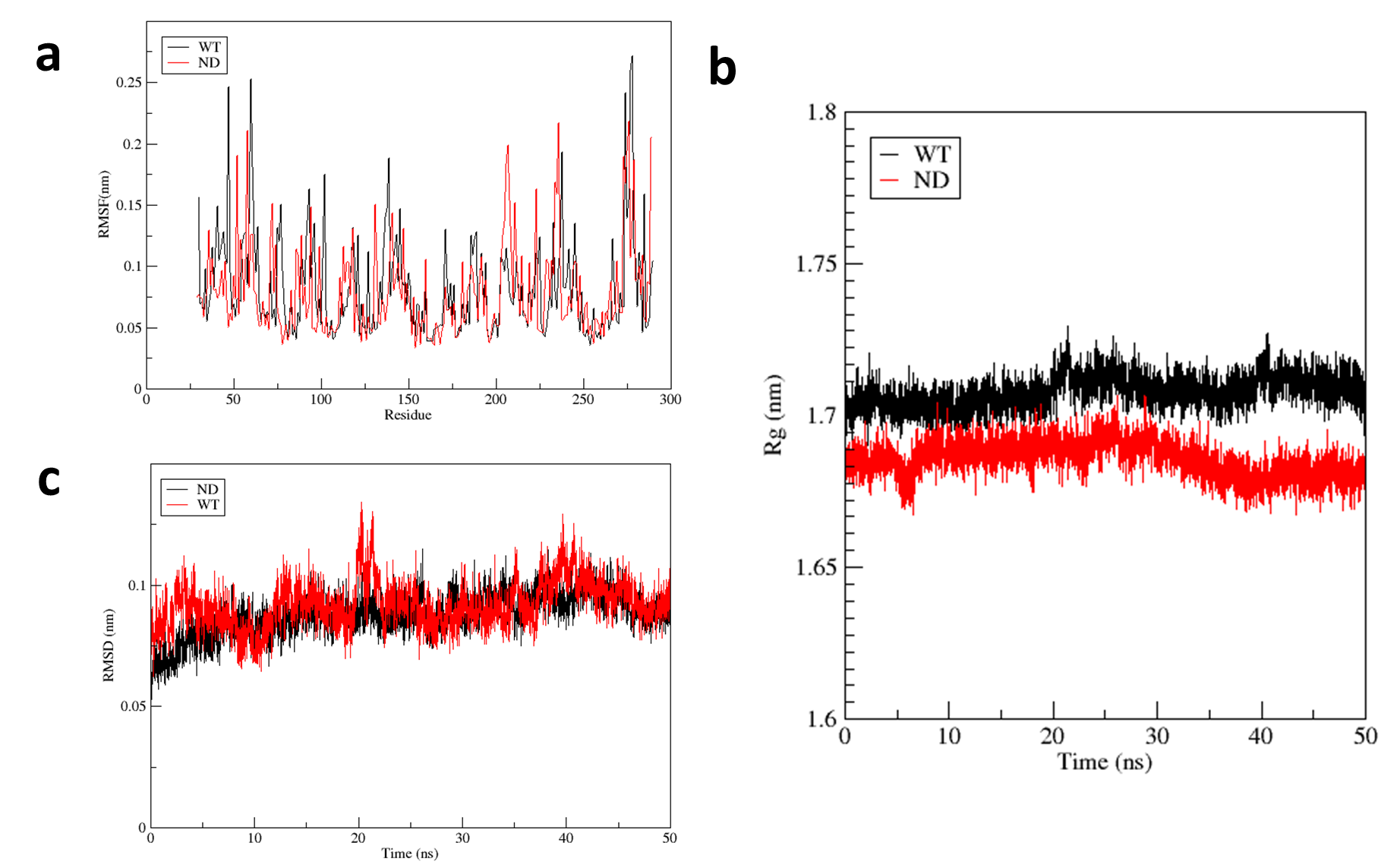
Figure S14. Molecular dynamics trajectories of IsPETase and ND were analyzed. IsPETase represents IsPETase and ND represents mutant ND. (a) The RMSF curve of the protein during the protein-ligand complex simulation process (The fluctuation of each atom relative to its average position is calculated, which characterizes the average of the structural changes over time, and gives a characterization of the flexibility of each region of the protein. The RMSF of the ND-ligand complex has a large difference from that of the IsPETase-ligand complex, indicating that the active region of the protein of the ND mutant shifted); (b) The Rg curve of the protein during the protein-ligand complex simulation process (The radius of gyration (Rg) is used to prove the compactness of the protein structure during the simulation process, it is the distance between the centroid of all atoms and their ends at a specific time interval. Throughout the complex MD simulation, the fluctuation of Rg value of IsPETase is larger than that of ND, indicating that ND binds more tightly to small molecules); (c) The RMSD curve of the protein during the protein-ligand complex simulation process (The complex reached equilibrium after 20 ns, indicating that the whole simulation process was stable and reliable. The RMSD of the ND composite system showed a small decrease compared to that of the IsPETase, indicating that the stability of the ND composite system was higher than that of the IsPETase).
TABLE
Table. 1S The primers which were used in this study.
|
Number |
Primers |
Nucleic acid sequences(5'to3') |
|
1 |
BslipA-R |
CAGTGGTGGTGGTGGTGGTGATTCGTATTCTGTCCTCCgcc |
|
2 |
BslipA-F |
GCAACATGTCTGCGCAGGCTGCTGAGCATAATCCAGTTGTGAT |
|
3 |
pHCM12M-SPC-F |
GGCGGAGGACAGAATACGAATCACCACCACCACCACCACT |
|
4 |
pHCM12M-SPC-R |
CACAACTGGATTATGCTCAGCAGCCTGCGCAGACATGTTG |
|
5 |
BslipA/IsPETase-Test-F1 |
CCGGGACTCAGGAGCATTTAA |
|
6 |
BslipA/IsPETase-Test-R1 |
TCTAGTAGAGAGCGTTCACCGACA |
|
7 |
16sRNA-F |
AGAGTTTGATCCTGGCTCAG |
|
8 |
16sRNA-R |
TACGGTTACCTTGTTACGACTT |
|
9 |
pHCM12M-PETase-R |
GTCTTGAGGCGCGCGGAAAGTTAGCCTGCGCAGACATGTTG |
|
10 |
pHCM12M-PETase-F |
GCGATTTTAGAACAGCAAATTGCTCACACCACCACCACCACCACT |
|
11 |
BslipA/PETase-F |
CGTTAACGTTAATCTTTACGATGGCGTTCAGCAACATGTCTGCGCAGGCT |
|
12 |
BslipA/PETase-R |
TCCGAAGATTGACTATTCTCAGATAAAGTTCAGTGGTGGTGGTGGTGGTG |
|
13 |
pHCM12M-BslipA/PETase-F |
CACCACCACCACCACCACTGAACTTTATCT |
|
14 |
pHCM12M-BslipA/PETase-R |
AGCCTGCGCAGACATGTTGCTGAACGCCA |
|
15 |
1028-M-F1 |
GTCCACGGTTATGGAGGGAATGATT |
|
16 |
1028-M-R1 |
AATCATTCCCTCCATAACCGTGGAC |
|
17 |
1028-M-F2 |
GAATGATTATAACTTCATCTCCCTTATG |
|
18 |
1028-M-R2 |
CATAAGGGAGATGAAGTTATAATCATTCCC |
|
19 |
1028-M-F3 |
AAGGAACGTATAGCGCAGTT |
|
20 |
1028-M-R3 |
AACTGCGCTATACGTTCCTT |
|
21 |
PETase-M-F1 |
TTCCGGGATATAACTCGAGACAGTCATCAATTA |
|
22 |
PETase-M-R1 |
TAATTGATGACTGTCTCGAGTTATATCCCGGAA |
Table S2. BsLipA (50μl) (Concentration system not using the (StarMut) error-prone PCR kit)
|
Reaction system(μl) |
1 |
2 |
3 |
4 |
5 |
|
2×Rapid Taq Master Mix |
25 |
25 |
25 |
25 |
25 |
|
Template DNA |
1 |
1 |
1 |
1 |
1 |
|
Primer 1(10 µM) |
2 |
2 |
2 |
2 |
2 |
|
Primer 2(10 µM) |
2 |
2 |
2 |
2 |
2 |
|
MgCl2 |
0.5 |
1 |
2 |
0.5 |
1 |
|
MnCl2 |
0.5 |
0.5 |
0.5 |
1 |
1 |
|
H2O |
18.5 |
18 |
17 |
17.5 |
17 |
Table S3. IsPETase(50μl)
|
Reaction system(μl) |
0 |
2 |
3 |
4 |
6 |
8 |
10 |
|
2×Rapid Taq Master Mix |
25 |
25 |
25 |
25 |
25 |
25 |
25 |
|
Template DNA |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
|
Primer 1(10 µM) |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
|
Primer 2(10 µM) |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
|
Enhancer |
0 |
2 |
3 |
4 |
6 |
8 |
10 |
|
H2O |
20 |
18 |
17 |
16 |
14 |
12 |
10 |
Table S4.IsPETase(50μl)
|
Reaction system(μl) |
1 |
2 |
3 |
4 |
5 |
6 |
|
2×Rapid Taq Master Mix |
25 |
25 |
25 |
25 |
25 |
25 |
|
Template DNA |
1 |
1 |
1 |
1 |
1 |
1 |
|
Primer 1(10 µM) |
2 |
2 |
2 |
2 |
2 |
2 |
|
Primer 2(10 µM) |
2 |
2 |
2 |
2 |
2 |
2 |
|
MgCl2 |
0.5 |
1 |
2 |
0.5 |
1 |
2 |
|
MnCl2 |
1 |
1 |
1 |
2 |
2 |
2 |
|
H2O |
18.5 |
18 |
17 |
17.5 |
17 |
16 |
Table S5. Strains which were used in this study.
|
Names |
Relevant characteristic |
Source |
|
SCK6 |
Em his nprE18 aprE3 eglSΔ102 bglT/bglS EV lacA::PxylA-comKv |
YanXin Laboratory, Nanjing Agricultural University |
|
KT2440 |
- |
YanXin Laboratory, Nanjing Agricultural University |
|
DH5α |
deoR endA1 gyrA96 hsdR17 (rk-mk+)recA1 relA1 supE44 thi- 1 Δ(lacZYA-argF)U169 Φ80lacZ ΔM15F - λ - |
Vazyme |
SCK6 Accession Number: The ultra-competent Bacillus subtilis strain SCK6 can be obtained from the Bacillus Genetic Stock Center (http://www.bgsc.org), with the accession number 1A976; KT2440 Accession Number: txid160488; pBBR-tph Accession Number: txid101510.
| Number | Name | Nucleic acid sequences (5' to 3') |
|---|---|---|
| 1 | The signal peptide sequence for extracellular expression in Bacillus subtilis |
ATGAGAAGTAAAAAATTGTGGATAAGT TTGTGTTTGCGTTAACGTTAATCTTTAC GATGGCGTTCAGCAACATGTCTGCGCAG GCT |
| 2 | The signal peptide sequence for expression in KT2440 | ATGAAATACCTGCTGCCGACCCTGCTG CTGGTCTGCTCCTCCTCGCTGCCCAGCC GGCGATGGCC |
| 3 | BsLipA | GCTGAGCATAATCCAGTTGTGATGGTG CACGGCATTGGTGGTGCCTCTTATAACT TTTTTTCTATTAAAAATTACTTGATTGG ACAAGGCTGGGATCGAAACCAATTATAT GCTATTGATTTCATAGACAAAACAGGAA ATAACCGCAACAATGGTCCGCGTCTATC GAGATTCGTCAAAGATGTGTTAGACAAA ACGGGTGCCAAAAAAGTAGATATTGTGG CTCATAGTATGGGCGGGGCGAACACGCT ATATTATATTAAGAATCTAGATGGCGGC GATAAAATTGAGAACGTTGTCACAATTG GTGGAGCAAACGGACTCGTTTCAAGCAG AGCATTACCAGGCACAGATCCAAATCAA AAAATTCTTTACACATCCGTCTATAGCT CAGCAGATCTTATTGTCGTCAACAGCCT CTCTCGTTTAATTGGCGCAAGAAACATC CTGATCCATGGCGTTGGCCATATCGGTC TATTAACCTCAAGCCAAGTGAAAGGGTAT ATTAAAGAAGGACTGAACGGCGGAGGAC AGAATACGAAT |
| 4 | IsPETase | CAAACGAACCCGTATGCGAGAGGCCCTA AATCCGACAGCCGCATCTCTTGAAGCAT CAGCAGGCCCGTTTACAGTTCGTTCTTT TACAGTTTCAAGACCGTCAGGATATGGA GCAGGAACAGTTTATTATCCGACAAATG CAGGCGGCACAGTTGGAGCAATTGCTAT TGTTCCGGGATATACAGCGAGACAGTCA TCAATTAAATGGTGGGGACCGAGATTGG CAAGCCATGGATTTGTTGTTATTACAAT TGATACGAACAGCACACTGGATCAACCG TCATCAAGATCTTCACAACAAATGGCAG CACTTCGTCAGGTTGCAAGCCTGAACGG CACGTCATCATCACCGATTTATGGCAAA GTTGATACAGCAAGAATGGGCGTCATGG GCTGGTCAATGGGCGGAGGAGGATCACT GATTTCAGCAGCAAATAATCCGTCACTT AAAGCAGCAGCGCCGCAAGCACCGTGGA TTCATCAACGAATTTTTCATCAGTTACA GTTCCGACGCTGATCTTTGCATGCGAAA ACGATAGCATTGCACCGGTGAACAGCAG CGCCCTGCCTATTTATGATTCAATGTCA AGAAATGCGAAACAATTTCTGGAAATTA ACGGCGGCTCACATTCTTGCGCAAATAG CGGAAATTCAAATCAGGCACTTATTGGC AAGAAGGGCGTTGCATGGATGAAAAGAT TTATGGATAATGATACGCGCTATAGCAC ATTTGCATGTGAAAATCCTAATAGCACG AGAGTGAGCGATTTTAGAACAGCAAATT GCTCA |
| 5 | ND | GCTAACCCTTATGAAAGAGGACCTAACC CTACAGATGCATTACTGGAAGCTCGCAG CGGACCTTTTTCAGTTTCAGAAGAGAAT GTGTCTAGACTTAGCGCTAGCGGATTTG GAGGCGGAACAATCTACTATCCTAGAGA AAACAATACGTACGGAGCAGTGGCAATT TCTCCTGGATATACGGGAACAGAAGCTA GCATTGCATGGCTTGGCGAACGCATTGC CTCTCACGGATTTGTTGTCATTACAATT GATACAATTACGACACTTGATCAACCGG ACTCTAGAGCCGAACAGTTAAATGCTGC ATTAAACCATATGATTAACAGAGCCTCA TCTACAGTGAGATCAAGAATCGATTCAT CTAGATTAGCAGTCATGGGACATAGCAT GGGAGGAGGAGGCTCTCTTCGCTTAGCT TCACAAAGACCGGACCTGAAAGCAGCAA TTCCTTTAACGCCGTGGCATCTGAACAA AACTGGAGCTCAGTGACAGTTCCGACGT TAATCATTGGCGCTGATTTAGATACAAT TGCCCCGGTCGCAACATCTGCCAAGCCT ATCTATAACTCTTTACCTAGCAGCATTA GCAAGGCATATCTTGAATTAGATGGAGC CACACATTTTGCGCCGAACATTCCTAACA AGATCATTGGCAAATATTCAGTAGCTTG GCTTAAGAGATTCGTCGACAATGACACA CGCTATACACAATTTCTTTGTCCGGGAC CGAGAGATGGATTATTTGGCGAAGTTGA AGAATACAGATCAACATGTCCTTTT |
| 6 | The result of fragment sequencing under the error-prone PCR system added 0 μL StarMut Enhancer | CAAACGAACCCGTATGCGAGAGGCCCTA AATCCGACAGCCGCATCTCTTGAAGCAT CAGCAGGCCCGTTTACAGTTCGTTCTTT TACAGTTTCAAGACCGTCAGGATATGGA GCAGGAACAGTTTATTATCCGACAAATG CAGGCGGCACAGTTGGAGCAATTGCTAT TGTTCCGGGATATACAGCGAGACAGTCA TCAATTAAATGGTGGGGACCGAGATTGG CAAGCCATGGATTTGTTGTTATTACAAT TGATACGAACAGCACACTGGATCAACCG TCATCAAGATCTTCACAACAAATGGCAG CACTTCGTCAGGTTGCAAGCCTGAACGG CACGTCATCATCACCGATTTATGGCAAA GTTGATACAGCAAGAATGGGCGTCATGG GCTGGTCAATGGGCGGAGGAGGATCACT GATTTCAGCAGCAAATAATCCGTCACTT AAAGCAGCAGCGCCGCAAGCACCGTGGA TTCATCAACGAATTTTTCATCAGTTACA GTTCCGACGCTGATCTTTGCATGCGAAA ACGATAGCATTGCACCGGTGAACAGCAG CGCCCTGCCTATTTATGATTCAATGTCA AGAAATGCGAAACAATTTCTGGAAATTA ACGGCGGCTCACATTCTTGCGCAAATAG CGGAAATTCAAATCAGGCACTTATTGGC AAGAAGGGCGTTGCATGGATGAAAAGAT TTATGGATAATGATACGCGCTATAGCAC ATTTGCATGTGAAAATCCTAATAGCACG AGAGTGAGCGATTTTAGAACAGCAAATT GCTCA |
| 7 | The result of fragment sequencing under the error-prone PCR system added 4 μL StarMut Enhancer | GATCACTGATTTGACGACGAAATAATCC GTCACTTAAAGCAGCAGCGCCGAAGC ACCGTGGGATTCATCAACGAATTTTCA TCAGTTACAGTTCCGAGCGTACTTTG CATGCGAACGGAACGATAGCATGCGC TGAACAGCAGCGCCGGCCTTATTATG TCAATGTCAAGAAATGGCGCGAACAA TTCTGGAAATTAACGCGGCGCTCAATTCT TGCAGCAATAGCGGAAATTCAATCAG GCACTTATTGGCAAGAAGGGCGTTGCA TGGATGAAAAGATTTATGGATAATGATG CGGCTATAGCACATTTGCATGTGAAAA TCCCTAATAGCACGAGAGTGAGCGATT AGAACAGCAAAATTGTCTCA |
| 8 | The result of fragment sequencing under the error-prone PCR system added 6 μL StarMut Enhancer | CAAACGAACCCGTATGCGAGAGGCCCT AATCCGACAGCCGCATCTCTTGAAGCA TCAGCAGGCCCGTTACAGTTCGTTCTTT TTACAGTTTCAAGACCGTCAGGATATGG AGCAGGAACAGTTTATTATCCGACAAAT GCAGGCGGCACAGTTGGAGCAATTGCT ATTGTTCCGGGATATACAGCGAGACAGT CATCAATTAAATGGTGGTGGGACCGAGA TTGGCAAGCCATGGATTTGTTGTTATTAC ATTGATACGAACAGCACACTGGATCAAC CGTCATCAAGATCTTCACAACAAATGG CACTTCGTCAGGTTGCAAGCCTGAACG CACGTCATCATCACCGATTTATGGCAAA GTTGATACAGCAAGAATGGGCGTCATGG GCTGGTCAATGGGCGGAGGAGGATCACT GATTTCAGCAGCAAATAATCCGTCACTT AAAGCAGCAGCGCCGCAAGCACCGTGG ATTCATCAACGAATTTTTCATCAGTTAC GTTCCGACGCTGATCTTTGCATGCGAAA ACGATAGCATTGCACCGGTGAACAGCAG CGCCCTGCCTATTTATGATTCAATGTCA AGAAATGCGAAACAATTTCTGGAAATTA ACGGCGGCTCACATTCTTGCGCAAATAG CGGAAATTCAAATCAGGCACTTATTGGC AAGAAGGGCGTTGCATGGATGAAAAGAT TTATGGATAATGATACGCGCTATAGCAC ATTTGCATGTGAAAATCCTAATAGCACG AGAGTGAGCGATTTTAGAACAGCAAATT GCTCA |
| 9 | The result of fragment sequencing under the error-prone PCR system added 8 μL StarMut Enhancer | CAAACGAACCCGTATGCGAGAGGCCCT AATCCGACAGCCGCATCTCTTGAAGCA TCAGCAGGCCCGTTACAGTTCGTTCTTT TTACAGTTTCAAGACCGTCAGGATATGG AGCAGGAACAGTTTATTATCCGACAAAT GCAGGCGGCACAGTTGGAGCAATTGCT ATTGTTCCGGGATATACAGCGAGACAGT CATCAATTAAATGGTGGTGGGACCGAGA TTGGCAAGCCATGGATTTGTTGTTATTAC ATTGATACGAACAGCACACTGGATCAAC CGTCATCAAGATCTTCACAACAAATGG CACTTCGTCAGGTTGCAAGCCTGAACG CACGTCATCATCACCGATTTATGGCAAA GTTGATACAGCAAGAATGGGCGTCATGG GCTGGTCAATGGGCGGAGGAGGATCACT GATTTCAGCAGCAAATAATCCGTCACTT AAAGCAGCAGCGCCGCAAGCACCGTGG ATTCATCAACGAATTTTTCATCAGTTAC GTTCCGACGCTGATCTTTGCATGCGAAA ACGATAGCATTGCACCGGTGAACAGCAG CGCCCTGCCTATTTATGATTCAATGTCA AGAAATGCGAAACAATTTCTGGAAATTA ACGGCGGCTCACATTCTTGCGCAAATAG CGGAAATTCAAATCAGGCACTTATTGGC AAGAAGGGCGTTGCATGGATGAAAAGAT TTATGGATAATGATACGCGCTATAGCAC ATTTGCATGTGAAAATCCTAATAGCACG AGAGTGAGCGATTTTAGAACAGCAAATT GCTCA |
| 10 | The result of fragment sequencing under the error-prone PCR system added 10 μL StarMut Enhancer | TCTGCTGCTTAGGAGATTTTCAGATGCA AATGTGCTATAGGCGCGTATCATTATCC AAATCTTTTCATCCATGACCAGCGCCTT TTGGCAAATAGTGCGCGATGGAATTTGC CGCTATTTGCCGAAGATTTGCTGGTCAA CGTTAATTCCGAAGATTGTGTGTGAGGG CTGACATTGAATCATAAATAGGGCAGGG CGGCTGTGTTTCACCGGTGCAATGCTAT CGTTTTCGCATGCAAAGATCAGGCTGGA ACTGTAACCTGATGAAAATGTGCTGGTG ATGAATCCCGGGTCGTTGGGGCTGGTGA CTGCTTAAGTGACCGGATTATTTGCTGC TGAACTAGTGATCCCTCCTCCGCATTTC GACCAGCCCAGGACGCCCATCTGCTGCA GTATCACGTTGCAATAAGGCTGGTGATG ATGAGTCCGTTGCTAGGTTGGCAGTGCA GACGAAGTGCCTCATTGTTGTGTCGAAA ATCTTGATGACGGTGTCAGTTGGTGTCT GTTGTGATCAATTGTAATAACAACAAAT CCATGGCCTGCCAATCTGCGGCTCCACC ATTTAGTGATGACTGCTGCTTGTCCAAT CCCGAACCAATAGCAATTGCTCCGAACT GTGCCGCTCGCATTGTGCGATAAACTGA CTGTTCGTGCTCCATATCCTGGACGTCT TGAACTGTAAAAGAACTGGTGACGTAAA CGGGCTGTGATGATGGCTGAGGATGACG GGGTTCGGTGGTGATGGTGCTGGTGGCG |
| 11 | The result of fragment sequencing under the error-prone PCR system added 0.5 μL Mg2+ and 1 μL Mn2+ (without using StarMut Error-Prone PCR Kit) | CTAACTGCCATCAGGCGCGCCTAAAA CAAACGAACCCGTATGCGAGAGGCCCT AATCCGACAGCCGCATCTCTTGAAGCA TCTGCAGGCCCGTTACAGTTCGTTCTCT TTACAGTTTCAAGACCGTCAGGATATGG AGCAGGAACAGTTTATTATCCGACAAAT GTAGGCGGCACAGTTGGAGCAATTGCT ATTGTTCCGGGATATACAGCGAGACAGT CATCAATTAAATGGTGGTGGGACCGAGA TTGGCAAGCCATGGATTTGTTGTTATTAC ATTGATACGAACAGCACACTGGATCAAC CGTCATCAAGATCTTCACAACAAATGG CACTTCGTCAGGTTGCAAGCCTGAACG CACGTCATCATCACCGATTTATGGCAAA GTTGATACAGCAAGAATGGGCGTCATGG GCTGGTCAATGGGCGGAGGAGGATCACT GATTTCAGCAGCAAATAATCCGTCACTT AAAGCAGCAGCGCCGCAAGCACCGTGG ATTCATCAACGAATTTTTCATCAGTTAC GTTCCGACGCTGATCTTTGCATGCGAAA ACGATAGCATTGCACCGGTGAACAGCAG CGCCCTGCCTATTTATGATTCAATGTCA AGAAATGCGAAACAATTTCTGGAAATTA ACGGCGGCTCACATTCTTGCGCAAATAG CGGAAATTCAAATCAGGCACTTATTGGC AAGAAGGGCGTTGCATGGATGAAAAGAT TTATGGATAATGATACGCGCTATAGCAC ATTTGCATGTGAAAATCCTAATAGCACG AGAGTGAGCGATTTTAGAACAGCAAATT GCTCA |
| 12 | The amino acid sequence of IsPETase |
MNFPRASRLMQAAVLGGLMAVSAAATA QTNPYARGPNPTAASLEASAGPFTVRSFT VSRPSGYGAGTVVYPTNAGGTVGAIAIV PGYTARQSSIKWWGPRLASGHFVVITIDT NSTLDQPSSRSSQQMAALRQVASLNGTS SSPJYGKVDTARMGVMGWMSGGGGSLLI SAANNPSLKAAAPQAPWDSSNTFSSVTV PTLIFACENDSIAPVNSSALPIYDSMSRNA KQFLEINGGSHSCANSGNSNQALIGKKG VAWMKRFMDNDTRYSTFACENPNSTRVS DFRTANCS |
| 13 | The amino acid sequence of TfCut |
MNFPRASRLMQAAVLGGLMAVSAAATA QTNPYARGPNPTAASLEASAGPFTVRSFT VSRPSGYGAGTVVYPTNAGGTVGAIAIV PGYTARQSSIKWWGPRLASGHFVVITIDT NSTLDQPSSRSSQQMAALRQVASLNGTS SSPJYGKVDTARMGVMGWMSGGGGSLLI SAANNPSLKAAAPQAPWDSSNTFSSVTV PTLIFACENDSIAPVNSSALPIYDSMSRNA KQFLEINGGSHSCANSGNSNQALIGKKG VAWMKRFMDNDTRYSTFACENPNSTRVS DFRTANCS |
| 14 | The amino acid sequence of ND |
ANPYERGPNPTDALLEARSGPFVSEEN VSRLSASGFGGGTIYYPRENNTYGAVAIS PGYTGEASIAWLGERIASGHFVVITIDTI TTLDQPDRAEQLNAALNHMINRASSTTV RSRIDSSRRLAVMGHSMGGGGGSLRLASQR PDLKAAIPLTPWHLNKNWSSSVTVPTLIIG ADLDTIAPVATSIAKPIYNSSLPSSISKAYLE LDGATHFAPNIPNKIIGKYSVAWLKRFVD NDTRYTQFLCPGPRDGFLGVEEEYRSRTC PF |