
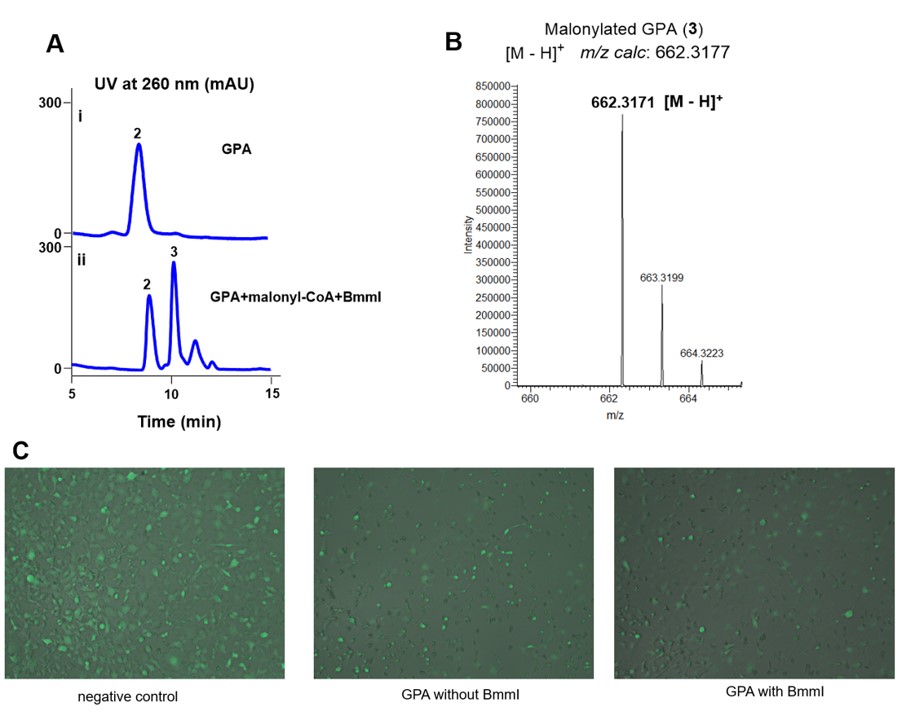
Previously, we identified BmmI could transfer malonyl group onto the anti-tumor compound GPA (Fig 1, 2). We optimized the reaction conditions, and found the convention rate could be ~50% in the presence of 20 μM enzyme (Fig 3A, 3B). To evaluate whether malonyl group could influence the cytotoxic activity of GPA, we thought to use fluorescent tumor cells to measure the cytotoxic activity by observing their density and morphology. We chose human fibrosarcoma cells HT1080 expressing the enhanced green fluorescent protein (EGFP), and respectively incubated it with a negative control without GPA, GPA without BmmI, and GPA with 20 μM of BmmI. After 24 hours, the density and morphology of the fluorescent cells were observed using fluorescence microscope. As shown in Fig 3C, compared to that of the negative control, the densities of the fluorescent tumor cells have reduced dramatically in the presence of GPA with or without BmmI, indicating both GPA and malonylated GPA have cytotoxic activity against HT1080. Furthermore, the density of fluorescent tumor cells in the presence of GPA and 20 μM BmmI (generating ~50% malonylated GPA) is much lower than that with the addition of GPA only. These results indicated that malonylation could improve the cytotoxic activity of GPA, prompting us to engineer BmmI by directed evolution for higher catalytic efficiency toward GPA.
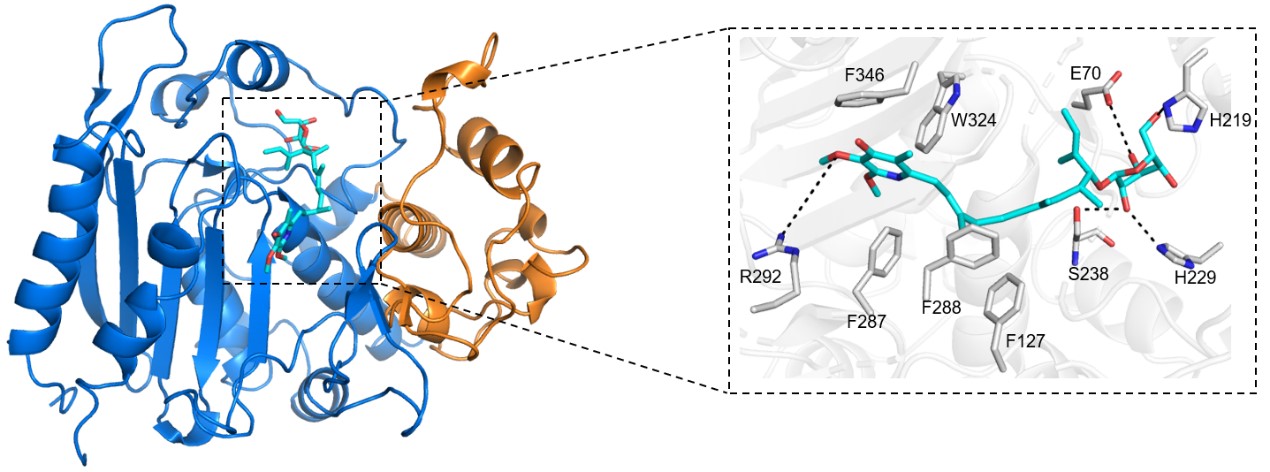
Previously we elucidated the crystal structure of BmmI (PDB id: 6KJH), which belongs to the structures of BLRE family enzymes [7]. To find out the possible binding residues of BmmI with GPA (2), we built the models for BmmI/GPA (2) through computer-aided molecular docking. As shown in Fig 4, GPA is located at the catalytic cavity of BmmI; the sugar moiety forms hydrogen bonds with Glu70, Ser238, His219 and His229, and the oxygen at the C3' of pyridine moiety hydrogen bonds with Arg292; in addition, the aliphatic chain of GPA is stabilized by nonpolar interactions with Phe346, Trp324, Phe287, Phe288 and Phe127. Based on the substrate binding mode, these residues were deduced to be potentially related to the catalytic activity of BmmI toward GPA. Therefore, we selected Glu70, Ser238, His219, His229, Arg292, Phe346, Trp324, Phe287, Phe288 and Phe127 for further directed evolution study.
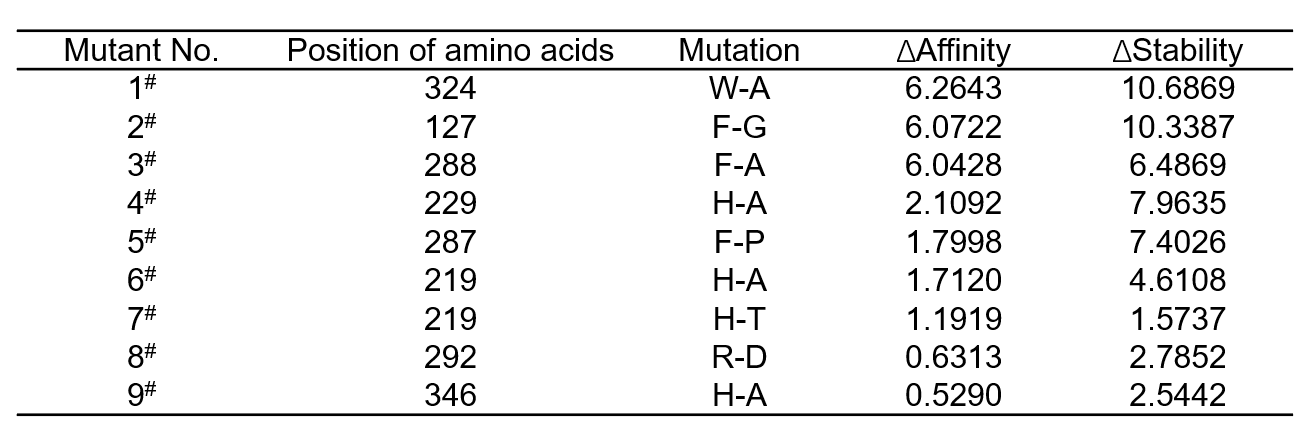
We constructed a virtual enzyme library of 190 saturated mutants of Glu70, Ser238, His219, His229, Arg292, Phe346, Trp324, Phe287, Phe288 and Phe127 using Molecular Operating Environment (MOE). To select the possible hints with higher catalytic efficiency, we performed docking-based virtual screening. We measured the binding affinities of these mutants with GPA, and their protein stabilities. As shown in Table 1, the top-ranking 9 mutants with high binding affinities toward GPA, and good stabilities were selected for further study.
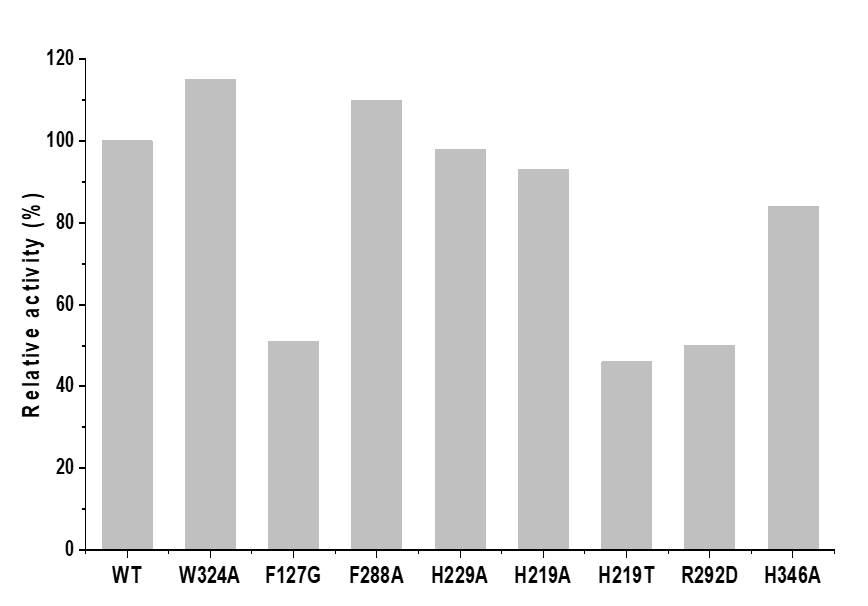
We then constructed the nine mutants W324A, F127G, F288A, H229A, F287P, H219A, H219T, R292D and H346A. Reverse complementary primers with mutation sites were designed to amplify the linear expression plasmids (pET28a/bmmI) with mutated sequences (Table S2). The resulting linear plasmids were ligated to circular molecules by seamless cloning, and then introduced into E. coli Rossetta (DE3). The expression strains were individually cultured, followed by addition of 0.2 mM IPTG to induce protein expression (Fig S1). Crude enzymes from each expression strain were incubated with GPA and malonyl-CoA for 2 h, and the generation of malonylated GPA was detected by HPLC. As shown in Fig 5, compared to that of the wild-type BmmI, the catalytic activities of W324A and F288A toward malonyl-CoA and GPA increased by ~15% and 10%.
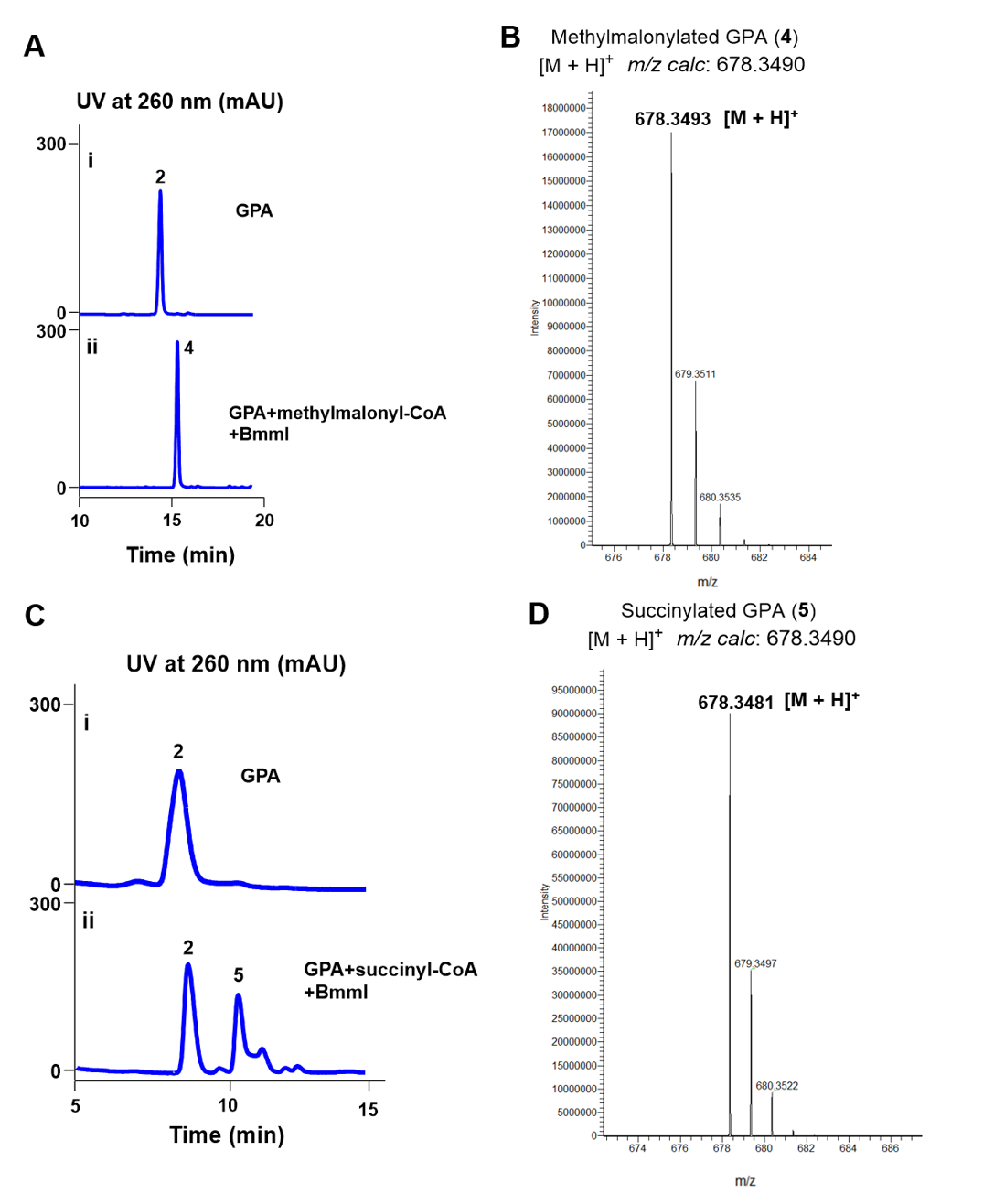
Furthermore, we tested the catalytic activities of W324A toward different acyl donors. We chose carboxyl-terminated methylmalonyl-CoA and succinyl-CoA as acyl donors. As shown in Fig 6, W324A could completely transform GPA (2) to methylmalonylated GPA (4, Fig 6A, 6B); and exhibited ~ 50% conversion rate to succinylated GPA (5, Fig 6C, 6D). These results indicated that the W324A mutant serves as an efficient enzyme for the malonylation/methylmalonylation/succinylation of GPA.
Acyltransfer is of crucial importance for the modification of many secondary metabolites. The Bacillus-derived acyltransferase BmmI could catalyze the malonylation of the anti-tumor agent GPA. In this project, we identified the malonyl group could improve the cytotoxic activity of GPA. By using MOE, we constructed a virtual enzyme library of 190 saturated mutants of the binding residues, and selected top9 mutants with highest binding affinities by structure-based virtual screening. The mutant W324A exhibited ~15% increased malonyltransfer activity compared to that of the wild-type BmmI, and exhibited high catalytic efficiency with methylmalonyl-CoA and succinyl-CoA. Given that malonylation could improve the cytotoxic activity of GPA, BmmI could be exploited as a tool enzyme in expanding the chemical diversity of these compounds with better bioactivities.