Our project aims to develop a dual selection system consisting of negative and positive selections to identify PobR mutants responsive to different aromatic compounds.
Biosensor
An allosteric TF (aTF), PobR
An aTF-based biosensor can respond to a specific compound, or a series of compounds with similar structures, and subsequently activate reporter gene expression [1-6]. The TF PobR belongs to the iclR superfamily, and is a positive transcriptional regulator of the pobA gene involved in 4-hydroxybenzoic acid (4HB) metabolism in Acinetobacter [7].
Last year, our team developed a PobR-based biosensor responsive to a 4HB analog, hydroxymandelic acid (HMA) [8]. These results strongly indicated that PobR had the potential to be modified to become biosensors for a variety of aromatic compounds.
Directed evolution
Construction of the PobR mutant library
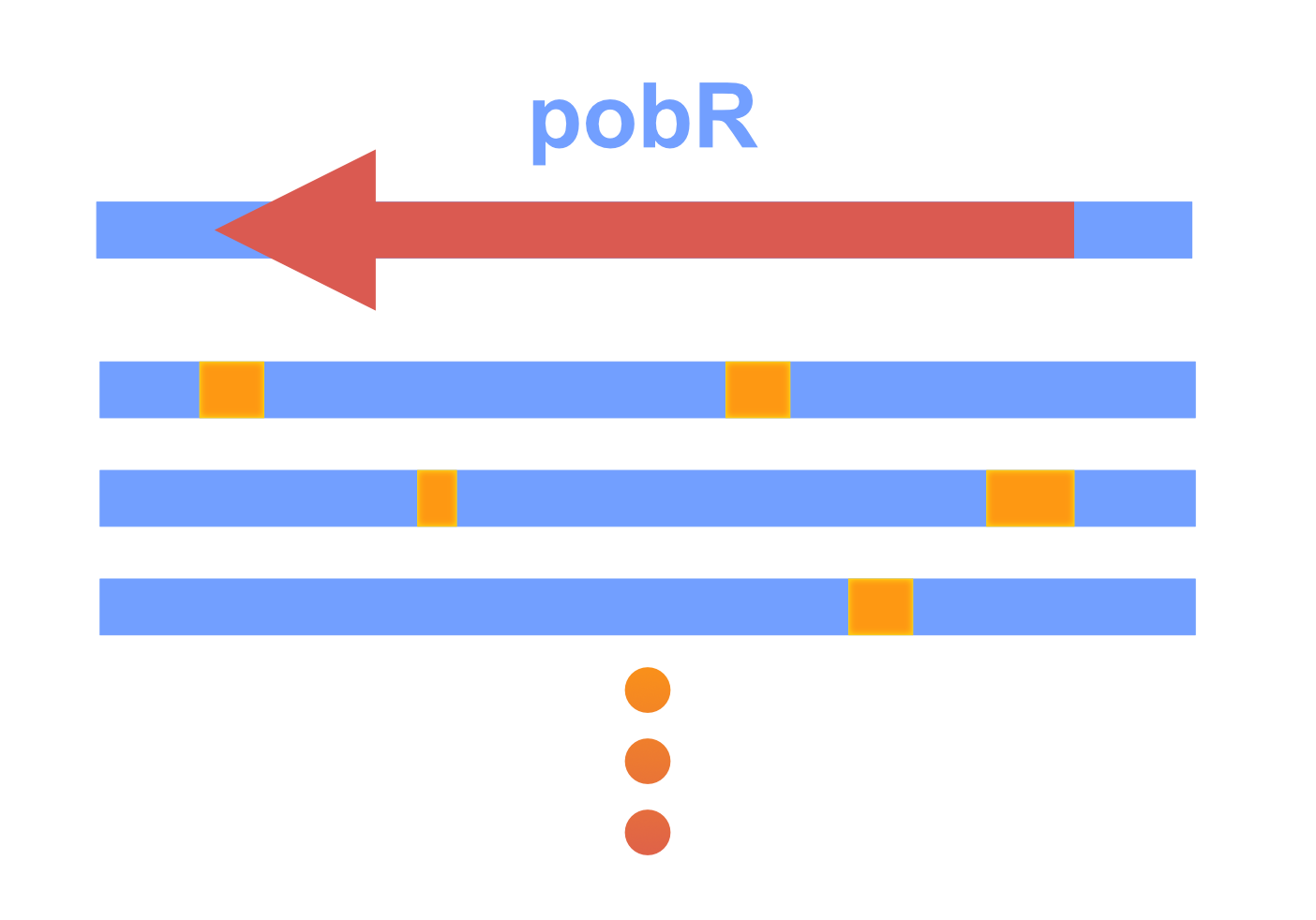
In order to construct a PobR library with random mutagenesis, we employed the approach of error-prone PCR amplification. The generated PobR mutants were subcloned into the vector containing PpobA, mCherry, codA and cmr.
The dual selection system
After random mutagenesis to generate a PobR library, we wanted to eliminate the PobR mutants that were either still responsive to 4HB or constitutively active in driving the PpobA independent of any ligand. For this purpose, we designed a dual selection system.
We first constructed a negative selection system consisting of the promoter PpobA and the CD enzyme coding sequence. The CD enzyme could convert exogenously added 5-FC to 5-FU that killed bacteria, and this mechanism allowed us to eliminate both PobR mutants still responsive to 4HB and pseudo-positive mutants.
Secondly, we constructed a positive selection system through inserting the chloramphenicol resistance gene downstream of the PpobA promoter to help us select the PobR mutants with desired ligand specificity.
We also added a reporter gene, mCherry, encoding a red fluorescence, in the selection system. The fluorescence intensity was proportional to the ligand binding affinity of PobR mutants that subsequently activated the PPobA promoter.
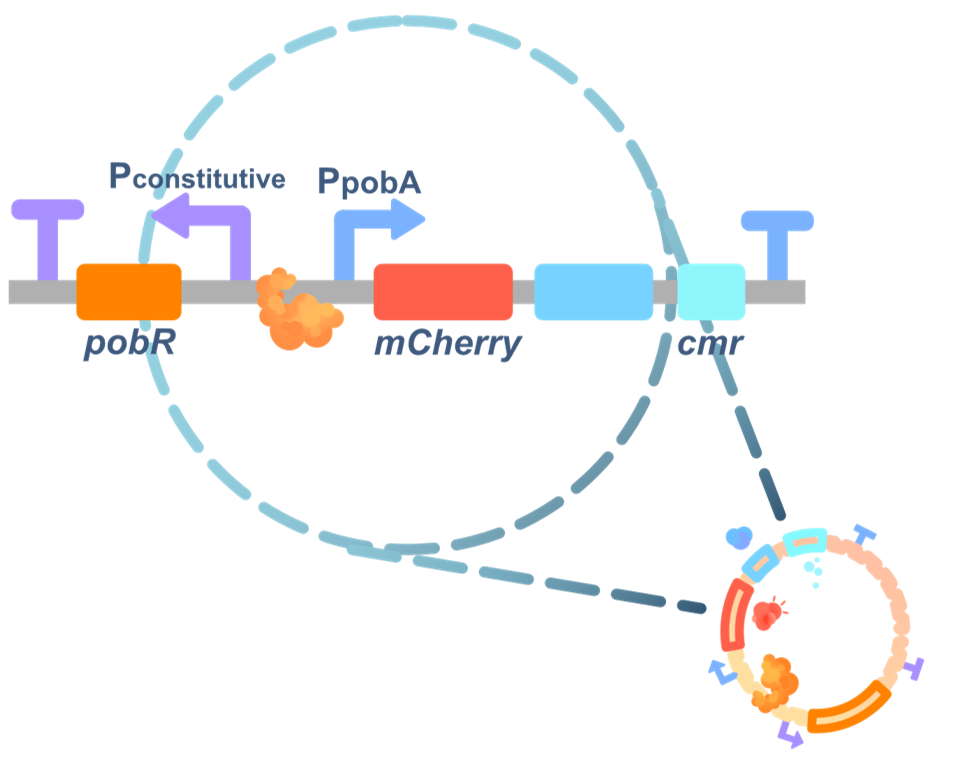
In the negative selection, the obtained bacteria were cultivated a liquid medium supplemented by 4HB and 5-FC.
In the positive selection, bacteria were cultured in a selective liquid medium supplemented with different aromatic compounds and chloramphenicol.
Next, we further characterized the responsiveness of selected PobR mutants to any aromatic compound through evaluating the induction of downstream genes and obtained mutants that were responsive to different aromatic compounds.

Test
Identification of amino acid mutations in selected PobR mutants
DNA sequencing of the selected PobR mutants was carried out to determine their mutation sites and classify the mutated amino acids.
Modeling and docking
To envision the effects of amino acid substitutions on the response of PobR proteins and their interactions with small molecule ligands, we used SWISS-MODEL [9] to simulate the protein structures and Autodock [10] to mimic the docking of different aromatic compounds to the proteins.
Using bioinformatic methods to process molecular docking, we evaluated how an amino acid mutation could affect ligand association with the binding pocket of the PobR mutant and predicted the alteration of binding affinity.
References
- Galvão TC, de Lorenzo V. Transcriptional regulators à la carte: engineering new effector specificities in bacterial regulatory proteins. Curr Opin Biotechnol. 2006; 17(1):34-42. doi: 10.1016/j.copbio.2005.12.002.
- Tang SY, Fazelinia H, Cirino PC. AraC regulatory protein mutants with altered effector specificity. J Am Chem Soc. 2008; 130(15):5267-71. doi: 10.1021/ja7109053.
- Taylor ND, Garruss AS, Moretti R, Chan S, Arbing MA, Cascio D, Rogers JK, Isaacs FJ, Kosuri S, Baker D, Fields S, Church GM, Raman S. Engineering an allosteric transcription factor to respond to new ligands. Nat Methods. 2016; 13(2):177-83. doi: 10.1038/nmeth.3696.
- Jha RK, Chakraborti S, Kern TL, Fox DT, Strauss CE. Rosetta comparative modeling for library design: Engineering alternative inducer specificity in a transcription factor. Proteins. 2015; 83(7):1327-40. doi: 10.1002/prot.24828.
- Jha RK, Kern TL, Kim Y, Tesar C, Jedrzejczak R, Joachimiak A, Strauss CE. A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor. Nucleic Acids Res. 2016; 44(17):8490-500. doi: 10.1093/nar/gkw687.
- Ellefson JW, Ledbetter MP, Ellington AD. Directed evolution of a synthetic phylogeny of programmable Trp repressors. Nat Chem Biol. 2018; 14(4):361-367. doi: 10.1038/s41589-018-0006-7.
- DiMarco AA, Averhoff B, Ornston LN. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J Bacteriol. 1993; 175(14):4499-506. doi: 10.1128/jb.175.14.4499-4506.1993.
- Liang Y, Luo J, Yang C, Guo S, Zhang B, Chen F, Su K, Zhang Y, Dong Y, Wang Z, Fu H, Sui G, Wang P. Directed evolution of the PobR allosteric transcription factor to generate a biosensor for 4-hydroxymandelic acid. World J Microbiol Biotechnol. 2022; 38(6): 104. doi: 10.1007/s11274-022-03286-5.
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46(W1):W296-W303. doi: 10.1093/nar/gky427.
- Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016; 11(5):905-19. doi: 10.1038/nprot.2016.051.
