Experimental Results
Results of Single-Site Saturation Mutagenesis
To evaluate whether single-site mutations could alter the enzyme’s DP specificity, we constructed a series of site-directed mutants including R159X, F170X, K172X, and R143X, and used un-purified mutated enzymes to catalyze polyM under identical conditions. To ensure comparability among different systems, the total product amount was adjusted to the same level before quantitative analysis depended on SDS-PAGE and TLC, followed by examination of the relative proportions of trisaccharide, tetrasaccharide, pentasaccharide, and hexasaccharide by HPLC.
Compared with the wild type PyAly, different mutants exhibited distinct product distributions. The tetrasaccharide proportion in the wild type was around 34.5%. At the K172 position, several mutants showed increased tetrasaccharide production, with K172D and K172E reaching the highest proportions of 42.1% and 41.4%, respectively, while K172W and K172Q also displayed elevated levels ranging from 36.3% to 38.4%.
At the R159 position, most mutations markedly increased tetrasaccharide content, with R159N, R159S, and R159H reaching 46.2%, 44.5%, and 42.4%, respectively, all well above the wild-type level. In addition, R159D, R159V, and R159L showed approximately 40% tetrasaccharide production (Figure 3,4).
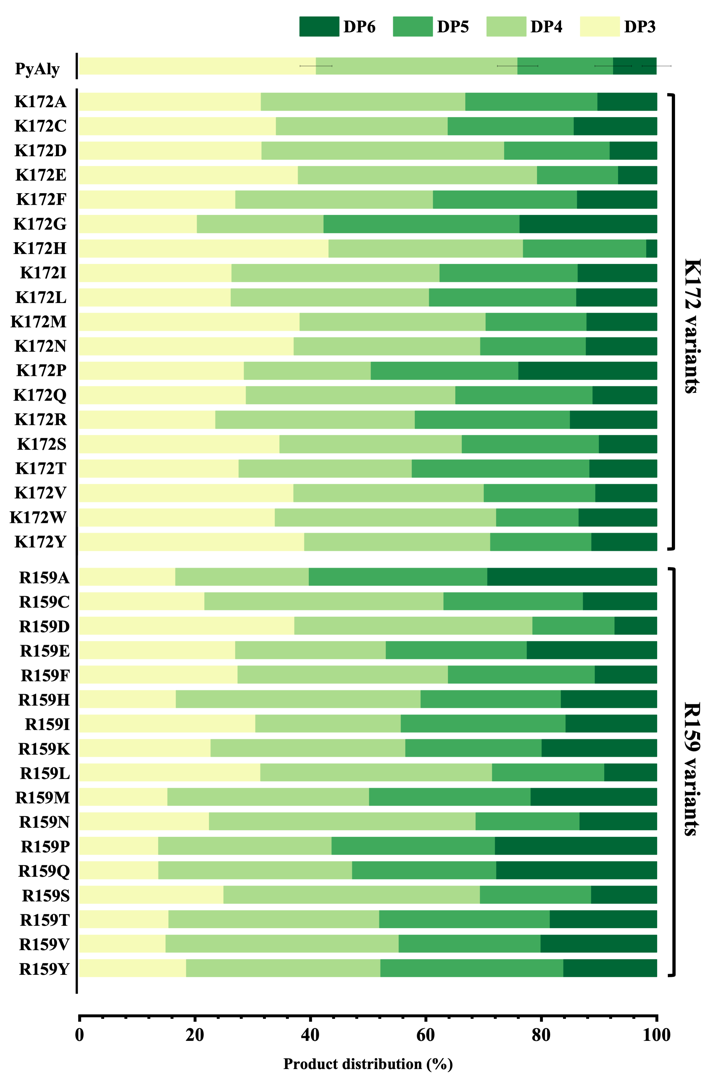
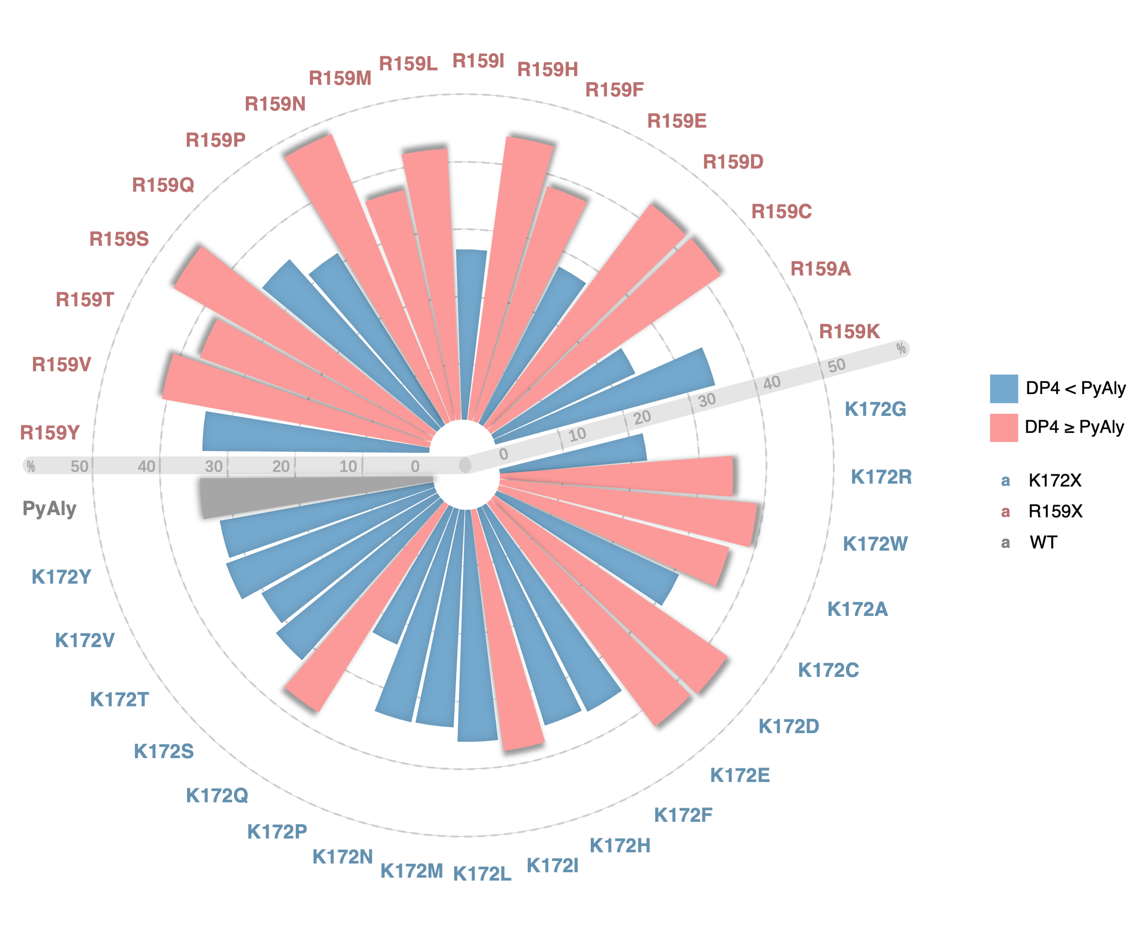
In summary, these results demonstrate that single-site mutations at different positions have distinct impacts on product distribution. Both the R159 and K172 series contain multiple “positive mutations” that increase tetra-saccharide proportion.
On the other sites, F170 was not included in this set of experiments, and R143 mutations exhibited relatively modest effects on tetrasaccharide proportion. Raw data are provided in Supplementary Materials 3.
Machine Learning Predictions for Double-Site Combinations
Considering the contributions of individual mutations to substrate binding in the “-” region, single-site data alone may be insufficient to predict double-mutant effects due to potential non-additive interactions. Therefore, a small set of double-site mutants serves as critical reference points to calibrate the machine-learning model and capture such interactions, improving prediction accuracy for untested variants.
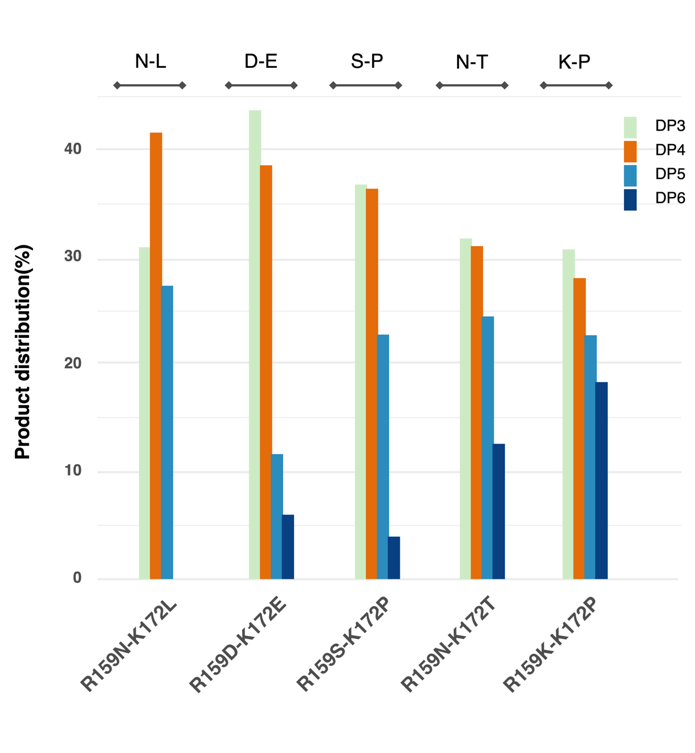
Based on this rationale, five double-site combinations were randomly selected to expand dataset coverage. These combinations (R159D-K172E, R159K-K172P, R159N-K172L, R159N-K172T, R159S-K172P) along with the wild type were constructed and characterized for their tetrasaccharide production ratios (DP4) and HPLC peak areas (Figure 5). This dataset, covering a broad range of activity and product selectivity, was used to train and vali-date the machine-learning model.
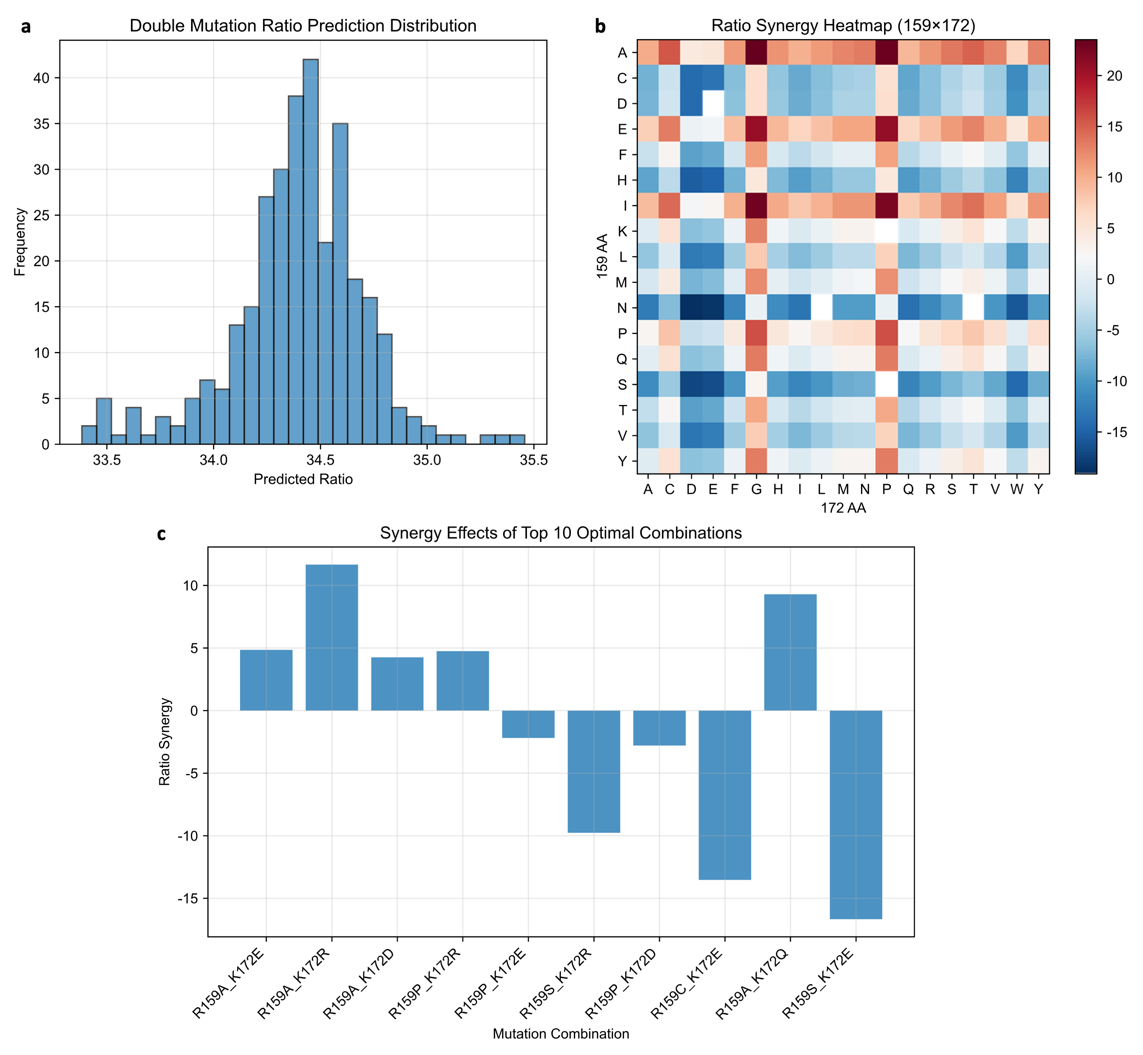
To investigate the combinatorial effects of R159 and K172 mutations, an esemble of machine-learning models—including SVR, MLP, GPR, and kernel-ridge regression—was used to exhaustively predict the performance of all 318 theoretical double-site variants (Figure 6). Both DP4 proportion and peak area were evaluated, and an integrated Combined Score was calculated to rank overall performance.
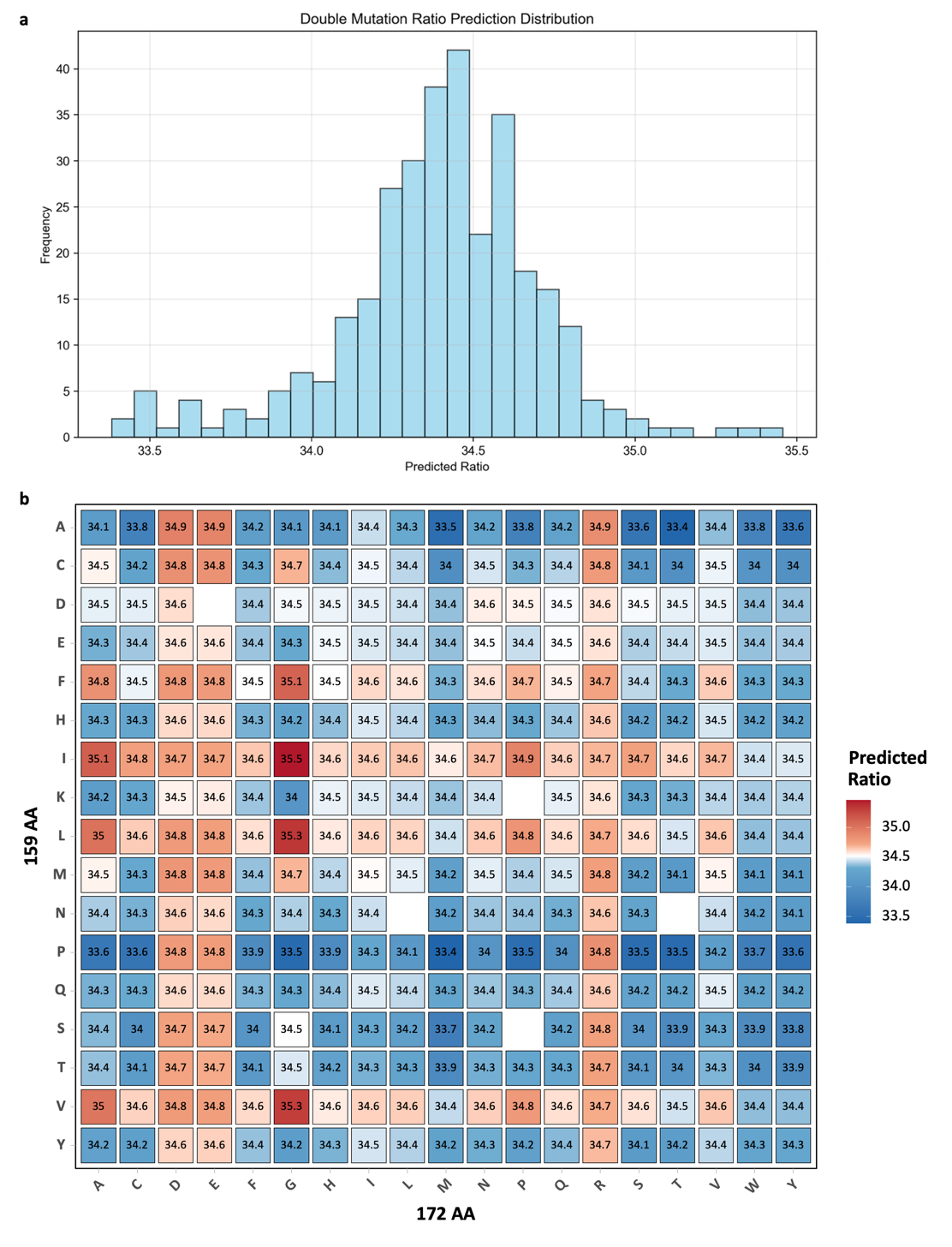
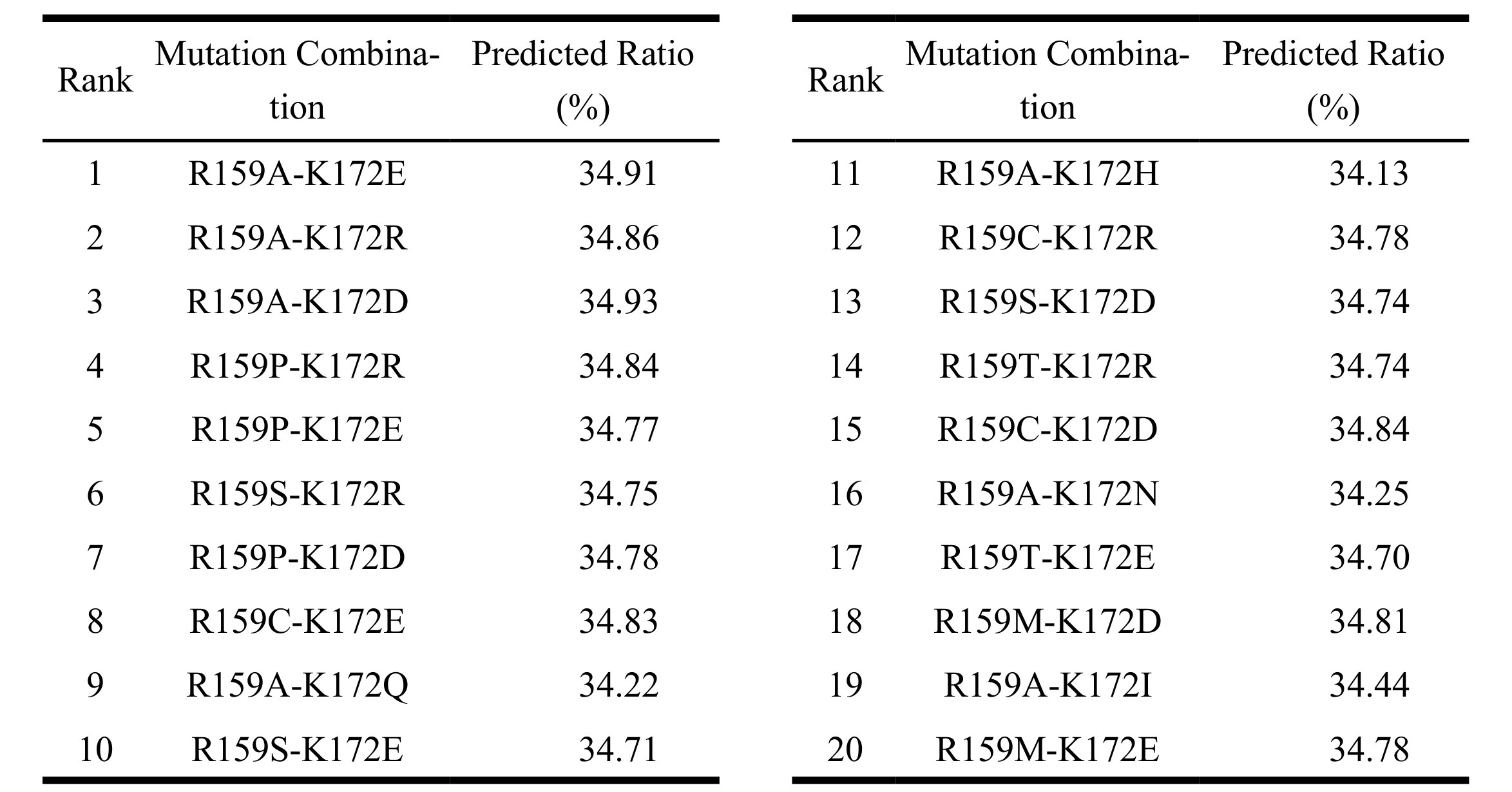
Among all candidates, the top twenty double-site mutants were predicted to achieve DP4 ratios ranging from 34.13% to 34.93%, mostly surpassing the wild-type PyAly (34.52%) (Table 1). The highest-ranked predicted combination was R159A-K172E, with a DP4 ratio of 34.91% and a Combined Score of 0.9882. Other top-performing predicted variants included R159A-K172R (34.86%, 0.9863) and R159A-K172D (34.93%, 0.9854). Variants with R159A/P substitutions paired with K172E/R/D consistently enhanced DP4 selectivity. Slightly lower-ranked variants (Combined Score ~0.973–0.979), such as R159A-K172H (34.13%, 0.9776) and R159C-K172R (34.78%, 0.9775), maintained DP4 ratios above 34.1%.
Overall, these predictions identify a set of promising double-site mutants for experimental evaluation.
Synergy Effect Analysis
The predicted tetrasaccharide proportions of all double-site variants are visualized in Figure 7, showing that most double mutants exhibit DP4 levels intermediate between their corresponding single mutants. Synergy analyses (Figure 7) quantify the difference between actual double-point effects and expected additive effects from single-point mutations. These results reveal that non-additive effects are generally minimal, indicating largely independent contributions of R159 and K172 to tetrasaccharide production. Moreover, explicit incorporation of such synergistic features into machine learning models improved predictive accuracy and facilitated the identification of top-performing double-point mutants.