
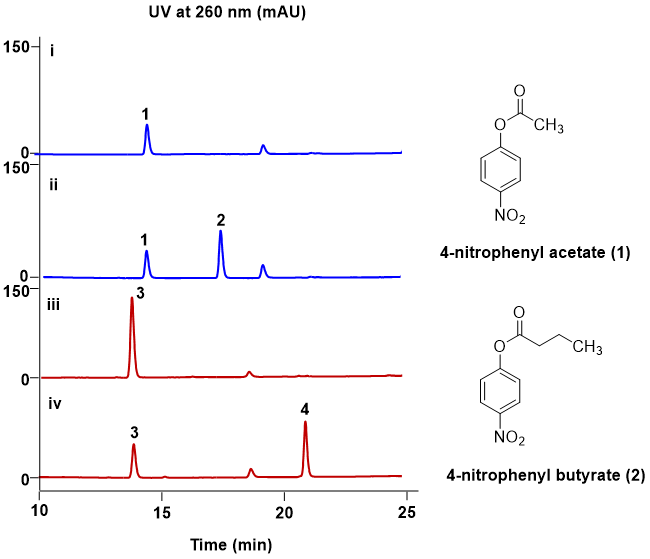
The P450BM3 gene was synthesized per the DNA sequence from GenBank (ID: KX768143.1). We then amplified the sequences encoding P450BM3-holoenzyme and P450BM3-heme domain, and cloned them into pET28a, respectively. The soluble form of P450BM3-holoenzyme (119 KDa) and P450BM3-heme domain (55 KDa) were obtained in Escherichia coli BL21 (DE3) (Fig S1). To find the possible substrates of P450BM3, we tested the catalytic activities of P450BM3 toward different small molecule compounds, including the benzene ring-containing compounds 3,4-dichloroaniline, 3,4-dichlorothiophenol, 4-nitrophenyl acetate, 4-nitrophenyl butyrate and 7-hydroxycoumarin, and the cholesterol-lowering compound lovastatin, anti-tumor compound epothilone D and the anti-oxidant compounds resveratrol and polydatin (Fig S2). The reactions were set up using P450BM3-holoenzyme (Heme-FMN-FAD) and NADPH as cofactor or P450BM3-heme domain and H2O2 as cofactor. As shown in Fig 2, in the presence of NADPH, P450BM3-holoenzyme (Heme-FMN-FAD) could transform 4-nitrophenyl acetate (1) to 3 as well as 4-nitrophenyl butyryl (2) to 4. Therefore, we choose 4-nitrophenyl acetate and 4-nitrophenyl butyryl as the substrates for P450BM3 for further study.
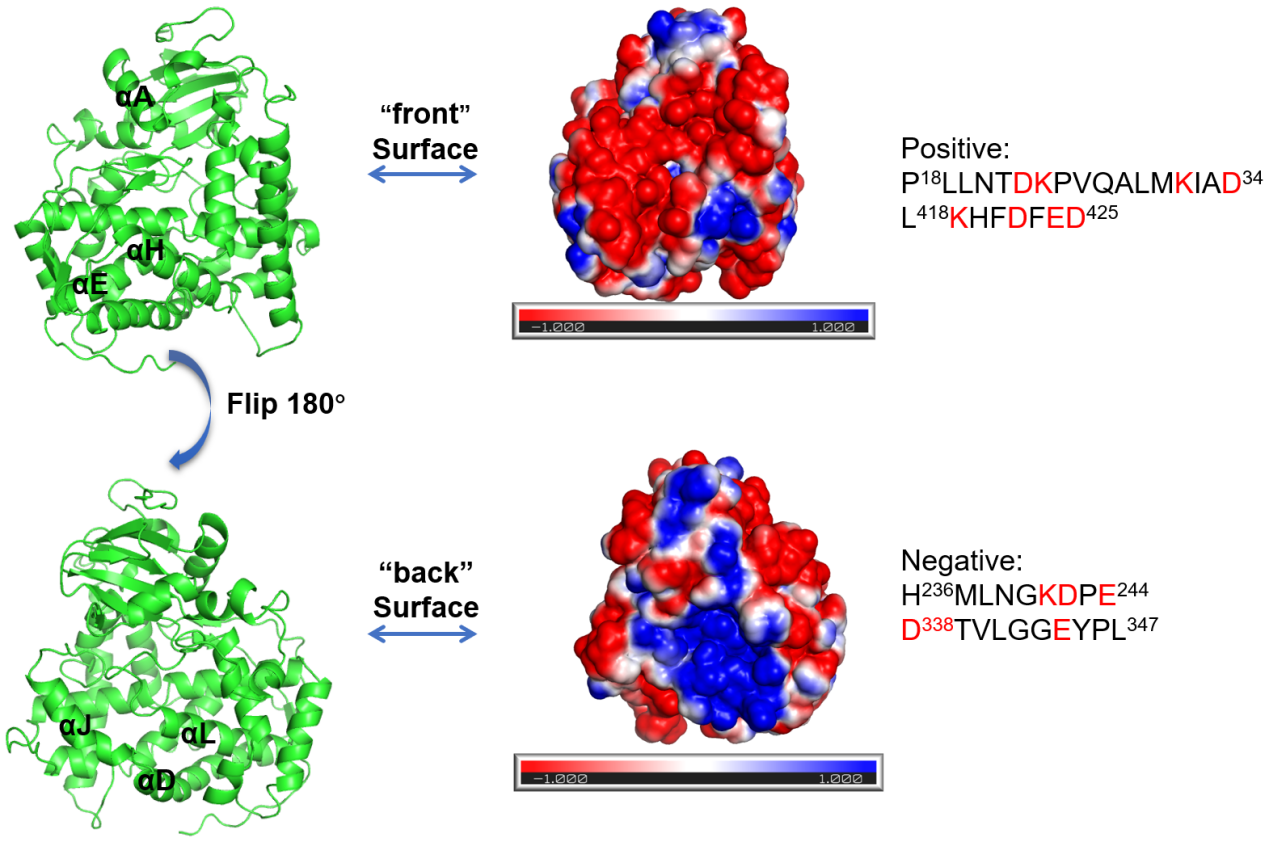
The crystal structure of P450BM3 was identified and deposited in PDB database (PDB id: 4ZFA). The protein surface was reported to be vital important for the enzyme to adapt to a particular environment. To probe the possible residues on the surface that may be closely related to the optimum pH of P450BM3, we used APBS to evaluate the electrostatic surface potentials of P450BM3. As shown in Fig 3, the “front” side of P450BM3 containing αA, αE and αH, is electronegative, while the “back” side of P450BM3 containing αD, αL and αJ, is electropositive. We then analyzed the amino acid sequences of these charged regions. “P18LLNTDKPVQALMKIAD34” and “L418KHFDFED425” constitute the negatively charged regions, and “H236MLNGKDPE244” and “D338TVLGGEYPL347” compose the positively charged regions. As it was reported that ionizable amino acids might have greater effect on pH optima than aliphatic ones, we choose the 13 amino acids with charged side chains (Asp23, Lys24, Lys31, Asp34, Lys419, Asp422, Glu424, Asp425, Lys241, Asp242, Glu244, Asp338 and Glu344) for further evolution study.
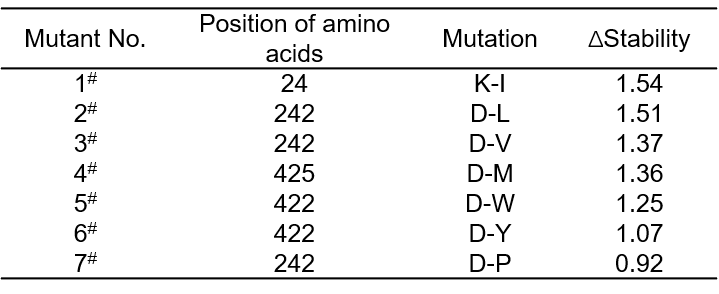
We constructed a virtual enzyme library of 247 saturated mutants of the 13 selected amino acids (Asp23, Lys24, Lys31, Asp34, Lys419, Asp422, Glu424, Asp425, Lys241, Asp242, Glu244, Asp338 and Glu344). To select the possible hints with higher stabilities, we used DynaMut website (https://biosig.lab.uq.edu.au/dynamut/) to measure the protein stability changes of these mutants. As shown in Table 1, the top-ranking 7 mutants with enhanced stabilities were selected for further study.
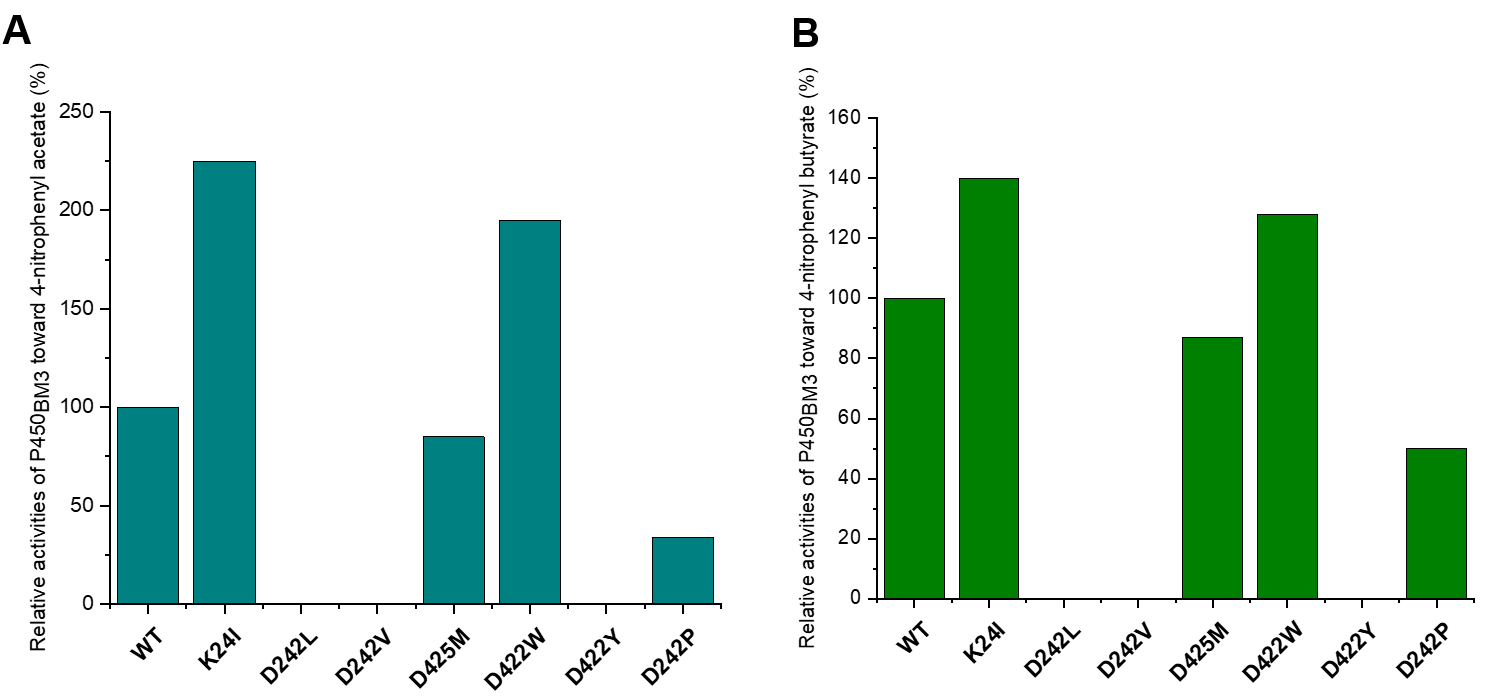
We then constructed the 7 mutants K24I, D242L, D242V, D425M, D422W, D422Y and D242P. Reverse complementary primers with mutation sites were designed to amplify the linear expression plasmids (pET28a/ P450BM3-holo) with mutated sequences (Table S2). The resulting linear plasmids were ligated to circular molecules by seamless cloning, and then introduced into E. coli BL21 (DE3). The expression strains were individually cultured, followed by addition of 0.2 mM IPTG to induce protein expression. Crude enzymes from each expression strain was incubated with NADPH and 4-nitrophenyl acetate or 4-nitrophenyl butyrate, respectively, for 2 h. The generation of products was detected by HPLC. As shown in Fig 4, compared to that of the wild-type P450BM3, the catalytic activities of K24I toward 4-nitrophenyl acetate and 4-nitrophenyl butyrate increased by ~1-fold and 0.4-fold, respectively; the catalytic activities of D422W toward 4-nitrophenyl acetate and 4-nitrophenyl butyrate increased by ~1-fold and 0.2-fold, respectively (Fig 4).
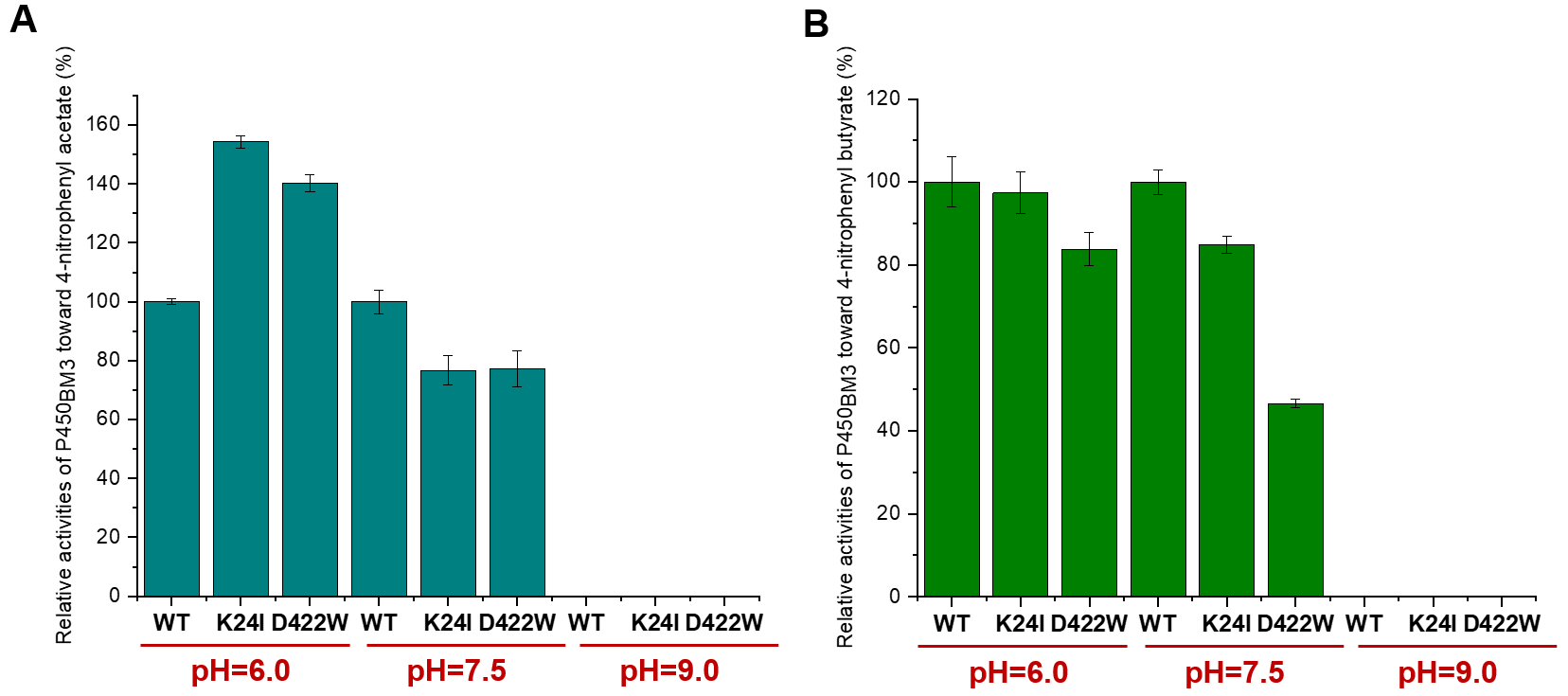
Furthermore, we purified the P450BM3 variants K24I and D423W (Fig S3), and tested the catalytic activities of the mutants and wild-type enzyme in acidic (Na2HPO3-Citric acid, pH=6.0), neutral (Na2HPO3-NaH2PO3, pH=7.5) and alkaline (Glycine-NaOH, pH=9.0) buffer, respectively. As shown in Fig 5A, in acidic buffer (pH=6.0), the catalytic activities of K24I and D422W increased by ~0.5-fold and 0.4-fold, respectively, compared to wild-type enzyme. These results indicated that the mutations of Lys24 and Asp422 could effectively improve the catalytic activities of P450BM3 in acidic conditions.
P450BM3 is a natural fusion enzyme comprising a heme-binding domain and a NADPH-P450 reductase domain. Its high activity and catalytic self-sufficiency make P450BM3 an excellent platform for biocatalysis. In this project, we analyzed the possible residues on the surface that may be closely related to the optimum pH of P450BM3 and constructed a virtual of 380 saturated mutants. The top7 mutants with enhanced stabilities were selected by using DynaMut website calculation. The variants K24I and D422W exhibited ~0.5-fold and 0.4-fold increased activities toward 4-nitrophenyl acetate in acidic condition (pH=6.0).