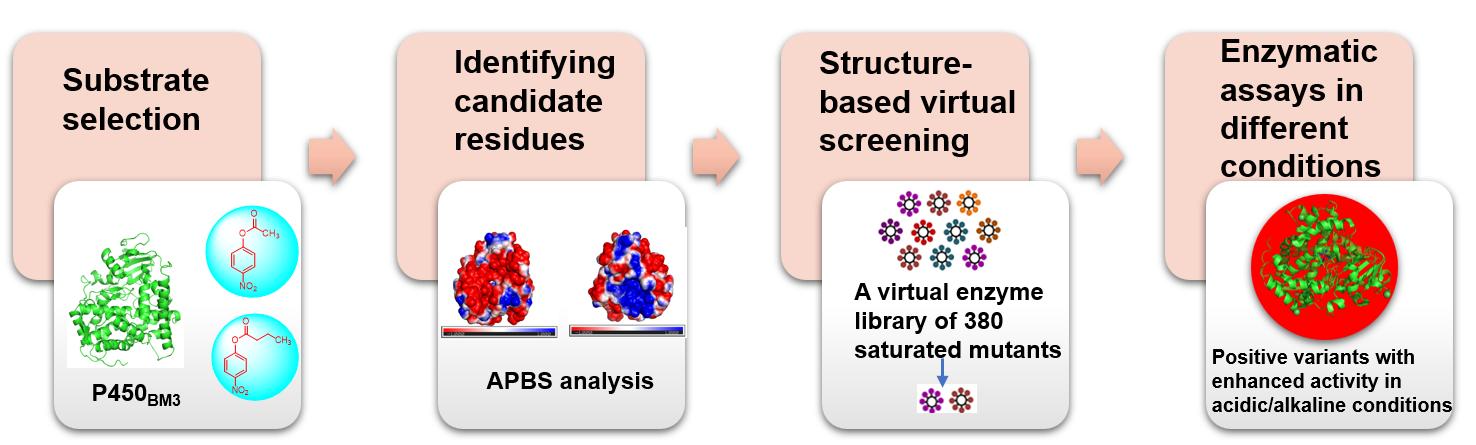
Cytochrome P450 enzymes (CYPs or P450s) are one of the most versatile biocatalyst systems in nature. P450s are capable of catalyzing more than 20 distinct types of reactions including hydroxylation, epoxidation, dealkylation, C−C bond cleavage, aromatic coupling, and others. Thus, P450s and their engineered variants are valuable tool enzymes in synthetic chemistry, synthetic biology, and green biomanufacturing. P450BM3 is a natural fusion enzyme in which a P450 heme domain (~55 kDa) is fused to an NADPH-P450 reductase (~65 kDa) domain through a flexible interdomain linker. It was first isolated from the soil bacterium Bacillus megaterium and was found to hydroxylate a range of different saturated fatty acids. As its high activity and catalytic self-sufficiency, P450BM3 has been engineered as a biotechnologically important enzyme with various new functions. For example, site-directed mutagenesis of P450BM3 removes its inherent substrate basis, enabling it to produce drug metabolites such as pravastatin, artemether, testosterone. P450BM3 has been engineered as an efficient peroxygenase by using dual-functional small molecules (DFSMs). Recently, P450BM3 variants that are able to catalyze intramolecular C-H amination were obtained by directed evolution. While, there were few studies about the modification of physicochemical properties of P450BM3. In practical terms, extreme processing conditions (such as high temperature, strong acid or alkali), severely impact the performance of natural enzyme, and restrict their application ranges. Therefore, in this study, we used semi-rational directed evolution to improve the stability of P450BM3 in acidic or alkaline conditions. We first evaluated the substrate selectivity of P450BM3. Then the possible amino acids relating to the stability of P450BM3 in acidic/alkaline conditions were analyzed. Furthermore, we used structure-based virtual screening to construct a saturated mutation library and selected suitable variants for expression in E. coli BL21 (DE3). Finally, the catal
Piericidins belong to a family of a-pyridone antibiotics, which feature a highly substituted pyridone core with a lipophilic side chain (Fig 1). Because of their structural resemblance to coenzyme Q10, piericidins have been demonstrated to be potent NADH-ubiquinone oxidoreductase (complex I) inhibitors in the mitochondrial electron transport chain. Among them, 10-glucopiericidin A1 (GPA, 2) is a lead compound with anti-renal cell carinoma efficacy (Fig 1). Considering that acyl groups often influence the bioactivities of natural products, in this article, we first evaluated the cytotoxic activity of malonylated GPA and found malonylation could improve the activity. Then the molecular docking studies were carried out to investigate the binding mode of GPA in BmmI. Furthermore, we used structure-based virtual screening to construct a saturated mutation library and selected suitable mutants for expression in E. coli Rossetta (DE3). Finally, mutants with enhanced malonylation activities were obtained and the catalytic activities of the mutant towards other acyl donors (methylmalonyl, succinyl) were assessed (Fig 2).