
freeze.yml and requirements.txt were provided in the folder "RetKcat", you can install the packages by running the following command in the folder you want to install RetKcat. The model that was trained on an RTX 3090 costs an average of 19 Gib memory and also 16 Gib on an RTX 4090 while predicting is available on an RTX 3070Ti with 6Gib on average via pre-trained model state provided by us. A large amount of evidence shows that this model will have a better performance with a higher hidden dimension, which is 64 in our work due to the limit of GPU memory.
First of all, run process.py in the child folder bin.
We recommend conda to construct an environment (may need to install some packages which were provided by others)
or try pip
try to download the code with git or zip
and run the code in the folder RetKcat,
put any sample you want to predict in the RetKcat/input.json and allow the format below
and run predict.py, out file will appear in the current folder, a temporary file input. pkl will be created and then removed after predicting.
The neural network can be divided into two parts: The first part utilizes a retentive network (RetNet) to extract protein features. This is achieved through a combination of causal masking and exponential decay along relative distances, which are combined into a single matrix. The second part employs graph convolutional networks (GCN) to capture substrate characteristics.
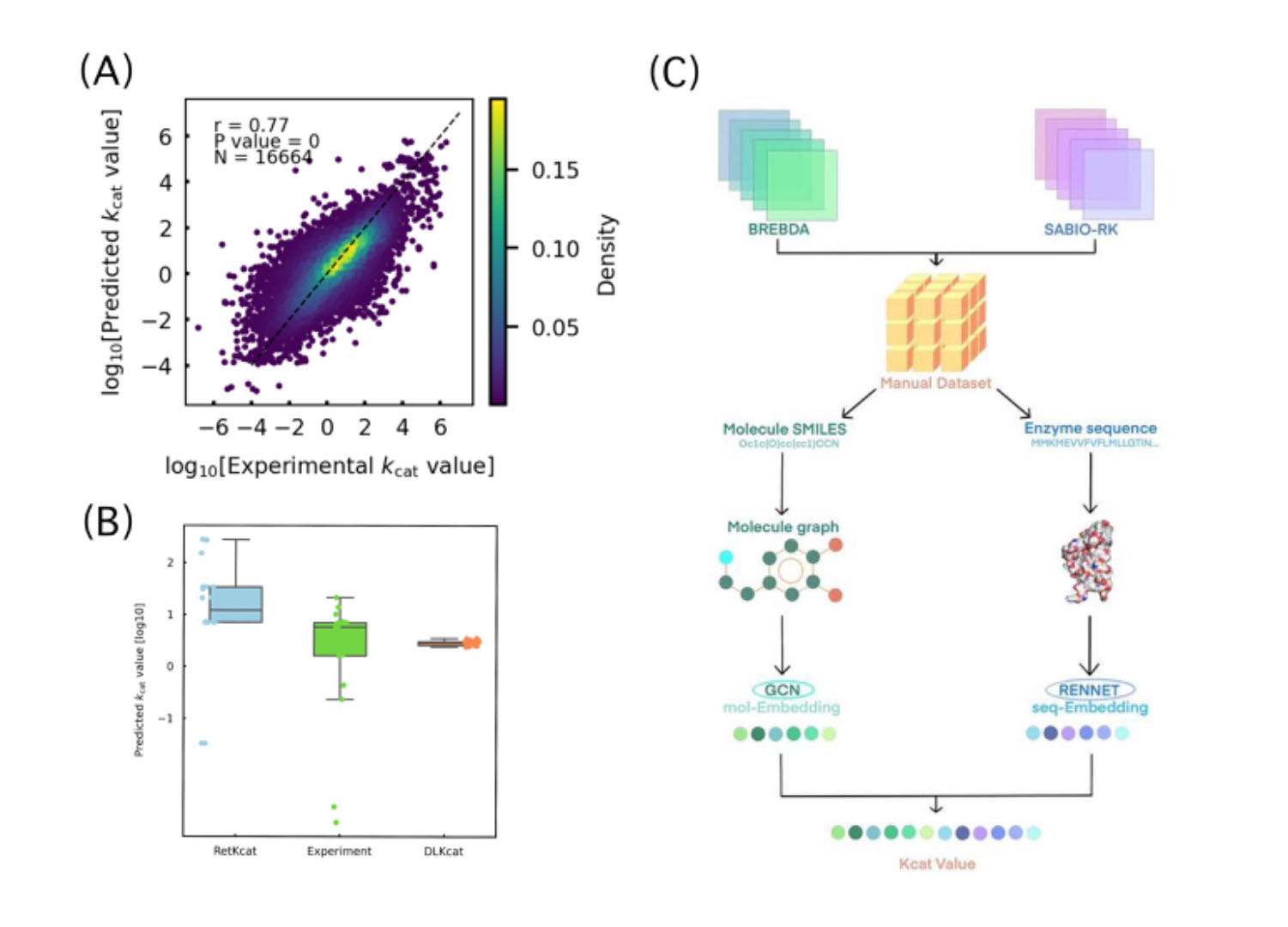
(A) RetKcat learning performance analysis. The trained model is tested on the training set, and R-square is used to measure whether the model has correctly learned the training set. (B) NCS samples prediction test. On the test set derived from the experiment, by comparing RetKcat with the currently better DLKcat. (C) RetKcat schematic diagram. RetKcat is composed of two parts, GCN is used to read molecular information, and RetNet is used to read protein information.
In this work, we developed an end-to-end learning approach for in vitro Kcat value prediction by combining a GCN for substrates and a RetNet for proteins. Molecular structures which are atoms linked with chemical bonds can be naturally converted into a graph and protein sequences can also be seen as a special format of the list.
First, substrate SMILES information was loaded with RDKit v.2022.9.5 (https://www.rdkit.org) and then each node will update itself via its neighbor around, which can be seen as dividing atoms with its chemical environment. Moreover, the adjacency of the molecule was extracted, and the molecule was finally represented as adjacency and an ordered node list. Then the edge information and node information have been convoluted. The final output of the GCN is a real-valued matrix M.
The protein sequence is manually split into ‘words’ which contain N amino acids. Every word is corresponding with a real number. Windows was set to limit the length of the word list, every N amino acid is transferred into numbers and held by Windows respectively. Then the matrix may be embedded to the appointed dimension. The protein representation and molecule representation will have the same dimension and will be concentrated as the input of RetNet.
The outcome of RetNet will forward an output layer, which consists of several Linear, and then the vector will be turned to predict value via a single layer Linear.
Databases including BRENDA (https://www.brenda-enzymes.org), and SABIO-RK (http://sabiork.h-its.org/) were used in the evaluation of the DLKcat performance. Protein Data Bank in Europe database (https://www.ebi.ac.uk/pdbe/).
Source data are provided in this paper. To facilitate further usage, we provide all codes and detailed instructions in the GitHub repository (https://github.com/CPU-CHINA/RetKcat). Files and results related to simulation calculations can also be found in the GitHub repository (https://github.com/CPU-CHINA/collation).
For more information: https://github.com/CPU-CHINA/RetKcat
1. Buller, A.R., et al., Directed evolution of the tryptophan synthase beta-subunit for stand-alone function recapitulates allosteric activation. Proc Natl Acad Sci U S A, 2015. 112(47): p. 14599-604.
2. Bunzel, H.A., J.L.R. Anderson, and A.J. Mulholland, Designing better enzymes: Insights from directed evolution. Curr Opin Struct Biol, 2021. 67: p. 212-218.
3. Cobb, R.E., R. Chao, and H. Zhao, Directed Evolution: Past, Present and Future. AIChE J, 2013. 59(5): p. 1432-1440.
4. Chan, S.K., et al., A semi-rational mutagenesis approach for improved substrate activity of microbial transglutaminase. Food Chem, 2023. 419: p. 136070.
5. Liu, Z., et al., Improvement of the acid resistance, catalytic efficiency, and thermostability of nattokinase by multisite-directed mutagenesis. Biotechnol Bioeng, 2019. 116(8): p. 1833-1843.
6. Amrein, B.A., et al., CADEE: Computer-Aided Directed Evolution of Enzymes. IUCrJ, 2017. 4(Pt 1): p. 50-64.
7. Jiang, L., et al., De novo computational design of retro-aldol enzymes. Science, 2008. 319(5868): p. 1387-91.
8. Li, D., et al., Improvement of catalytic activity of sorbose dehydrogenase for deoxynivalenol degradation by rational design. Food Chem, 2023. 423: p. 136274.
9. Li, R., et al., Computational redesign of enzymes for regio- and enantioselective hydroamination. Nat Chem Biol, 2018. 14(7): p. 664-670.
10. Liu, L., S. Zhou, and Y. Deng, Rational Design of the Substrate Tunnel of β-Ketothiolase Reveals a Local Cationic Domain Modulated Rule that Improves the Efficiency of Claisen Condensation. ACS Catalysis, 2023. 13(12): p. 8183-8194.
11. Duan, B. and Y. Sun, Integration of Machine Learning Improves the Prediction Accuracy of Molecular Modelling for M. jannaschii Tyrosyl-tRNA Synthetase Substrate Specificity. 2020.
12. Li, F., et al., Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction. Nature Catalysis, 2022. 5(8): p. 662-672.
13. Gado, Japheth E., Matthew Knotts, Ada Y. Shaw, Debora Marks, Nicholas P. Gauthier, Chris Sander, and Gregg T. Beckham. "Deep learning prediction of enzyme optimum pH." bioRxiv (2023). doi: https://doi.org/10.1101/2023.06.22.544776.
14. Mazurenko, S., Z. Prokop, and J. Damborsky, Machine Learning in Enzyme Engineering. ACS Catalysis, 2019. 10(2): p. 1210-1223.
15. Saito, Y., et al., Machine-Learning-Guided Mutagenesis for Directed Evolution of Fluorescent Proteins. ACS Synth Biol, 2018. 7(9): p. 2014-2022.
16. Yang, K.K., Z. Wu, and F.H. Arnold, Machine-learning-guided directed evolution for protein engineering. Nat Methods, 2019. 16(8): p. 687-694.
17. Wu, S., et al., Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew Chem Int Ed Engl, 2021. 60(1): p. 88-119.
18. Chen, X., et al., Photoenzymatic Hydrosulfonylation for the Stereoselective Synthesis of Chiral Sulfones. Angew Chem Int Ed Engl, 2023. 62(23): p. e202218140.
19. He, Y., et al., Discovery and Engineering of the l-Threonine Aldolase from Neptunomonas marine for the Efficient Synthesis of β-Hydroxy-α-amino Acids via C–C Formation. ACS Catalysis, 2023. 13(11): p. 7210-7220.
20. Herger, M., et al., Synthesis of beta-Branched Tryptophan Analogues Using an Engineered Subunit of Tryptophan Synthase. J Am Chem Soc, 2016. 138(27): p. 8388-91.
CPU_CHINA hosted The 1st Forum on International Directed Evolution Competition (iDEC) China on August 26.
In this exchange meeting, we invited iDEC teams from universities in China to participate in this event. The presenters shared the teams' latest research progress, analysis results and development trends in the field of directed evolution. In the process of this exchange and learning, everyone has gained a lot, and the friendship between the teams has been further deepened.
Thank you for your sharing and wish you all a good performance in the coming competition!

Poster of The ist Forum on iDEC China
© 2023 CPU_CHINA, 639 Longmian Avenue, Nanjing, Jiangsu, China