Background
Tyrian purple, mainly composed by 6,6'-dibromoindigo (6BrIG), is a dye extracted from the murex shellfish which was first produced by the Phoenician city of Tyre in the Bronze Age. It then spread in popularity and was adopted by the Romans as a symbol of imperial authority and status. The glands of thousands of putrefied crushed shellfish left to bake in the sun, with the overwhelming smell from the process. The resulting liquid was used to dye cloth fibres in manipulated variations of colours ranging from pink to violet. To obtain 1.4g tyrian purple, 12,000 Mediterranean snails would be killed. Its difficulty of manufacture, striking purple to red colour range, and resistance to fading made clothing dyed using Tyrian purple highly desirable and expensive.
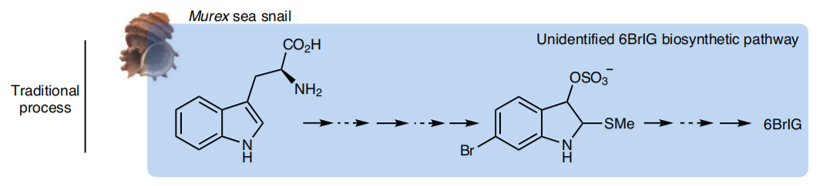
For chemical synthesis, there are still no productive methods known for industrialization. This challenge often comes from the difficulty of introducing the two bromine atoms in 6BrIG.
First, most methods use bromine gas or hydrogen bromide for bromination, but they suffer from lack of regiospecificity, low product yield and environmentally toxic manufacturing processes. Second, when synthesis is started from brominated precursors, the high cost of these precursors becomes an obstacle for large-scale synthesis.
Therefore, the bromination process is still too inefficient and uneconomical to be feasible on an industrial scale for either biological or chemical synthesis of 6BrIG.
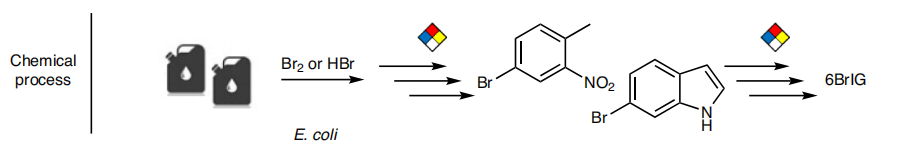
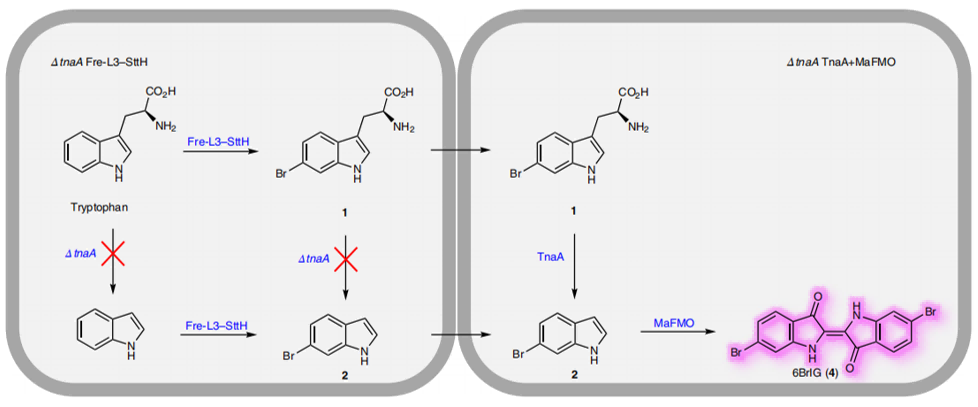
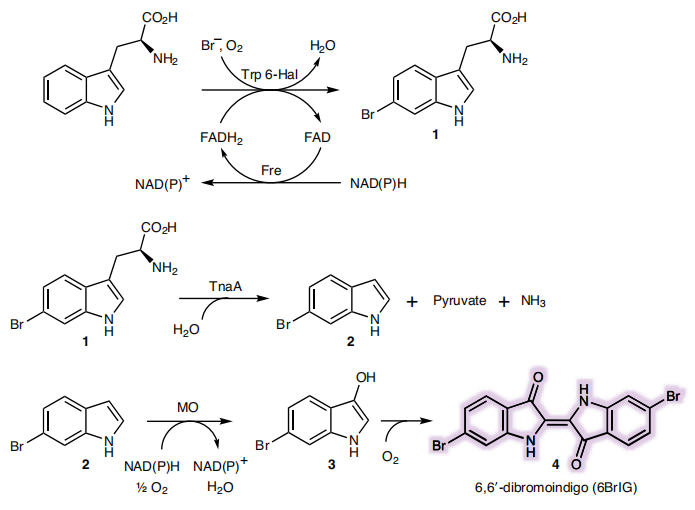
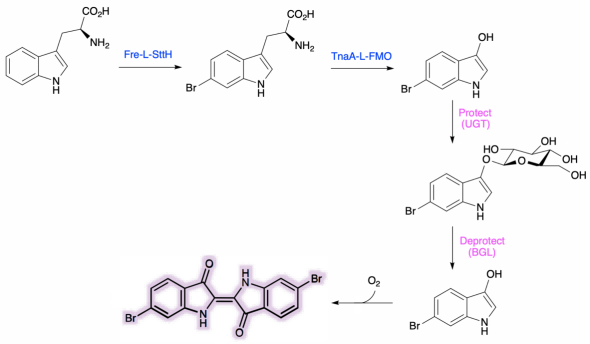
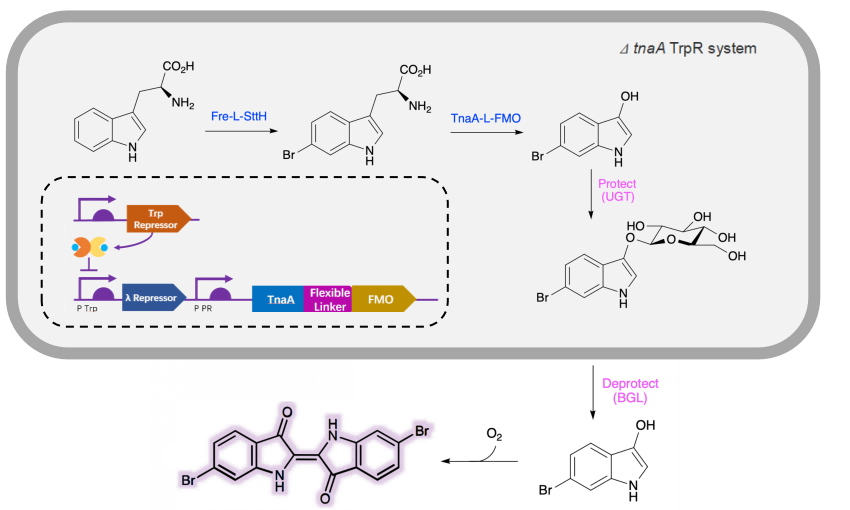
As a green and convenient production is preferred, syntheitc biology could be a better choice. Lee et al reported the first production strategy of Tyrian purple in E.coli using an accessible substrate tryptophan (Trp). Lee used three enzymes Trp 6-halogenase (Trp 6-Hal), tryptophanase (TnaA) and monooxygenase (MO) to finish this multiple-stepproduction.
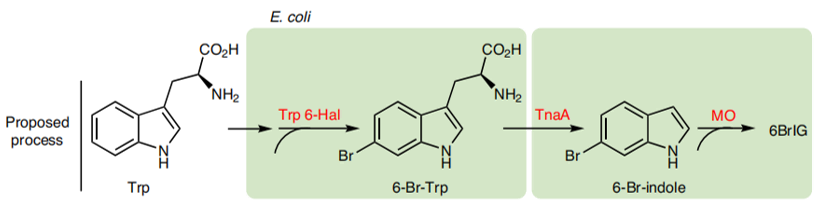
In addition, a consecutive two-cell system is designed as a solution to the basic problem of byproduct indigo in tyrian purple. TnaA can react with both Trp and the first step product Br-Trp. So the first step using Fre-SttH and second step using TnaA have to be in two different strains. To prevent the effect of endogenous TnaA expression, a ΔtnaA strain of E. coli was used and TnaA was overexpressed using a plasmid when needed throughout this study. Thus, a ΔTnaA E. coli strain without endogenous TnaA expression and another ΔTnaA E. coli strain with extra TnaA expression are constructed to form a two-cell system for production of tyrian purple.