Result dry Lab
Rational design the mutants
Results from Jennifer Doudna's group indicated that the α7 helix of the CasΦ protein has an important effect on enzymatic activity. From this, we modified the protein using a point mutation. The detailed mutation content is shown in Table 1. Mut-1 and Mut-2 are designed by Doudna’s group and used as positive controls. Mut-3 to Mut-6 are mutants designed by our team.
Table 1. Mutation sites of different mutants
Molecular dynamics simulation for mutants
We used the optimized system to carry out molecular dynamics simulation for the above mutants. According to the gmx rms module in the Gromacs package, we calculate the RMSD of different components in the simulation system. We analyzed the changes of six mutants in the system, and analyze the changes in the skeleton carbon atom of the protein and the central structure of the entire protein.
The average structure after 400 ns of the simulation was extracted to check the RMSD and Rg of the simulated trajectory respectively. As shown in Figure 1, in the two mutant types of CasΦ (Mut-1 and Mut-4), the RMSD for the whole system is far away, with a large shift after 100 ns, after which it keeps converging. While the RMSD of the wild-type CasΦ and other mutants present the stability of about 0.32 nm after 150 ns, with less vibration of the system. This stability result may be related to the conformation changes induced by hydrogen bonds within the protein molecule.
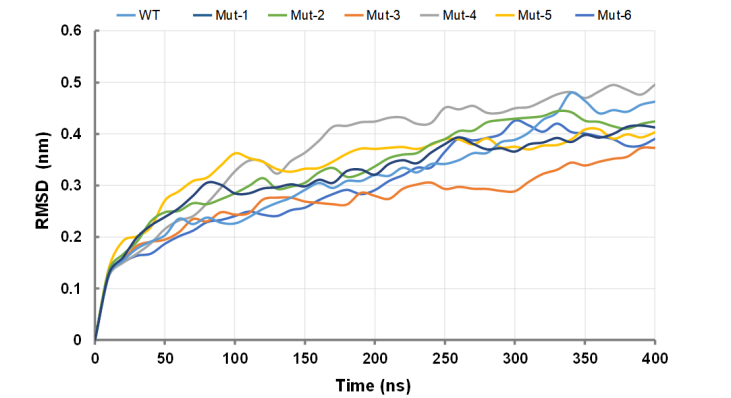
Figure1. Average RMSD value for wild-type CasΦ and mutants.
As shown in the Figure 2, after 400 ns of folding, the molecular structure of the seven proteins has partially changed. After 350 ns, they begin to converge and maintain a relatively stable fluctuation range. Among them, the conformational changes of Mut-3 and Mut-4 after a period of equilibrium before 200 ns are small, the convergence degree is high, and the system stability is good. The wild-type protein has a large structural change at 60 and 150 ns, and the internal interaction force is unstable after three-dimensional folding. In the positive control group (Mut-1 and Mut-2), the basic fluctuation range was small after 100 ns, and some small fluctuations occurred at 3.66 nm.
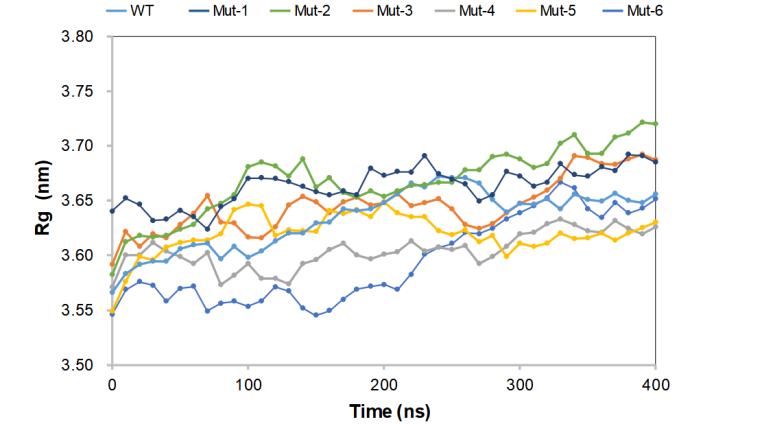
Figure 2. Average Rg value for wild-type CasΦ and mutants.
As shown in Figure 3, in the simulated 400 ns, the energy fluctuation amplitude of the whole system is small, the wild-type protein (WT) is the lowest, and the structural stability is the best, followed by the stability of Mut-2 and Mut-3, and the energy of the whole system remains unchanged.
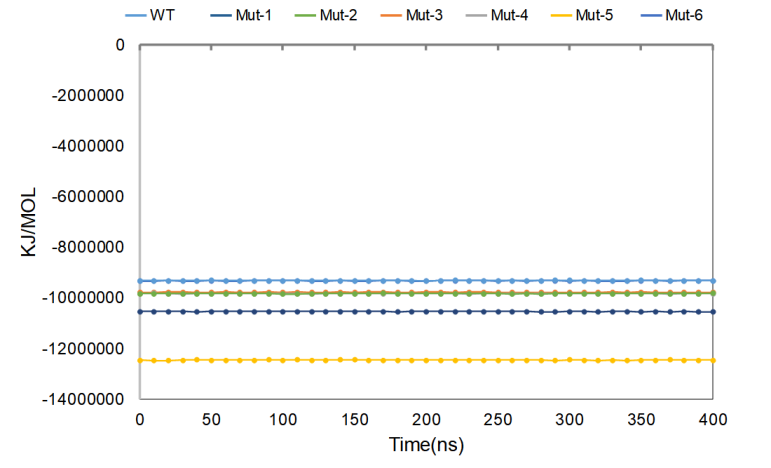
Figure 3. Average Gromacs energies value for wild-type CasΦ and mutants.
During the simulation process, the number of intramolecular hydrogen bonds in each group of proteins fluctuated to a certain extent, and changed at about 490. Among them, the vibration amplitude of Mut-4 and Mut-6 is smaller than that of other proteins, and the number of hydrogen bonds in the system has been maintained all the time, indicating that the intramolecular interaction is relatively stable, and it is easy to maintain a more rigid protein conformation. (Figure 4)
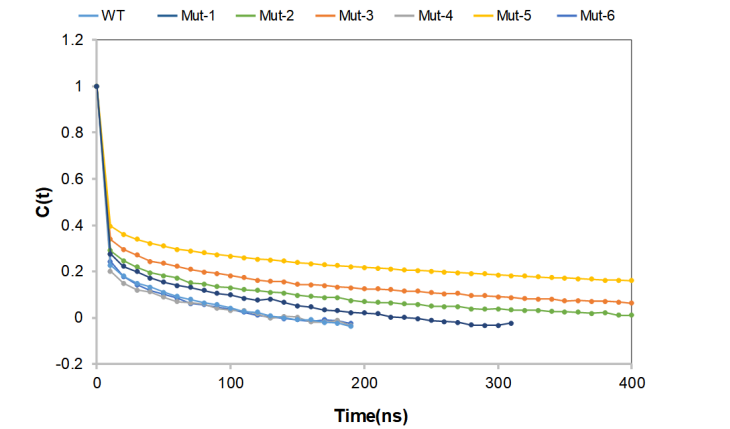
Figure 4. Intramolecular hydrogen bonds value for wild-type CasΦ and mutants.
During the simulation, the displacement range of each atom is small, and it is basically stable. In the Mut-3 and Mut-5 protein groups, the amino acids at 202-218 shifted by 1 nm, indicating that the intramolecular force has changed. The positive control group Mut-1 and Mut-2 basically maintained the displacement below 0.5 nm, and the atomic vibration amplitude was small, indicating that the system was stable. (Figure 5)
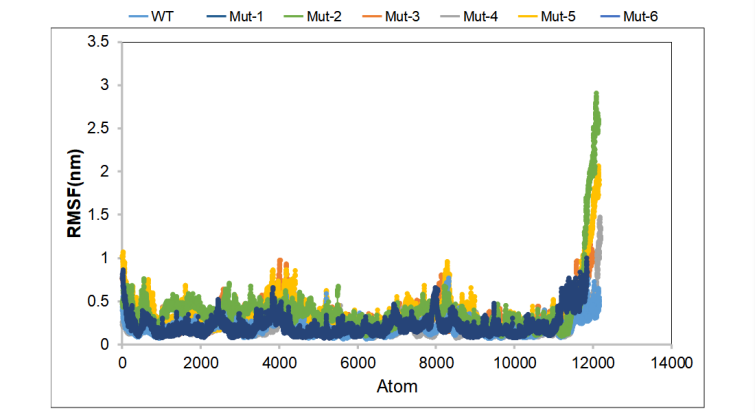
Figure 5. Average RMSF value for wild-type CasΦ and mutants.
Reference:
[1]Huyke, Diego A et al. “Enzyme Kinetics and Detector Sensitivity Determine Limits of Detection of Amplification-Free CRISPR-Cas12 and CRISPR-Cas13 Diagnostics.” Analytical chemistry vol. 94,27 (2022): 9826-9834. doi:10.1021/acs.analchem.2c01670
[2] Ramachandran, Ashwin, and Juan G Santiago. “CRISPR Enzyme Kinetics for Molecular Diagnostics.” Analytical chemistry vol. 93,20 (2021): 7456-7464. doi:10.1021/acs.analchem.1c00525
[3]Nguyen, Giang T et al. “Miniature CRISPR-Cas12 endonucleases - Programmed DNA targeting in a smaller package.” Current opinion in structural biology, vol. 77 102466. 25 Sep. 2022, doi:10.1016/j.sbi.2022.102466