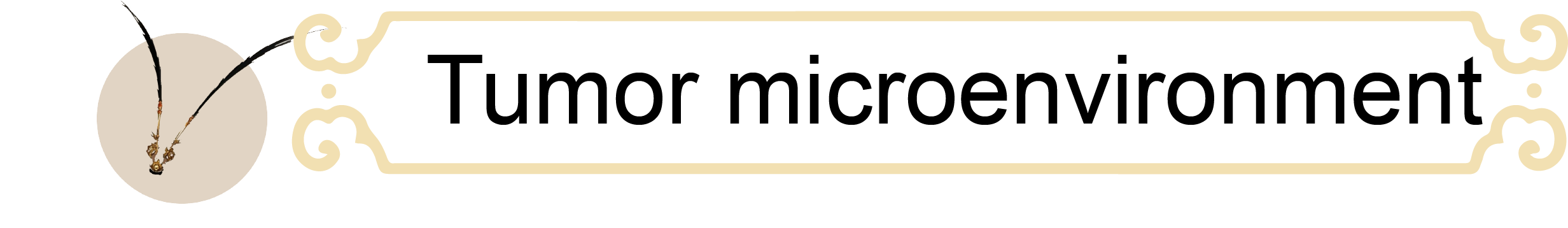
Adoptive cell therapy (ACT) has revolutionized clinical treatment strategies; however, it still faces considerable challenges in the context of solid tumors. These limitations are largely attributed to the immunosuppressive tumor microenvironment (TME), characterized by acidity, physical barriers, and overexpression of inhibitory checkpoints, which collectively restrict infiltration and induce exhaustion and dysfunction of T cells and natural killer (NK) cells. Moreover, cytokines essential for immune activation, particularly interleukin-2 (IL-2), are scarce within the TME. Together, these factors culminate in effector cell exhaustion and impaired activation.

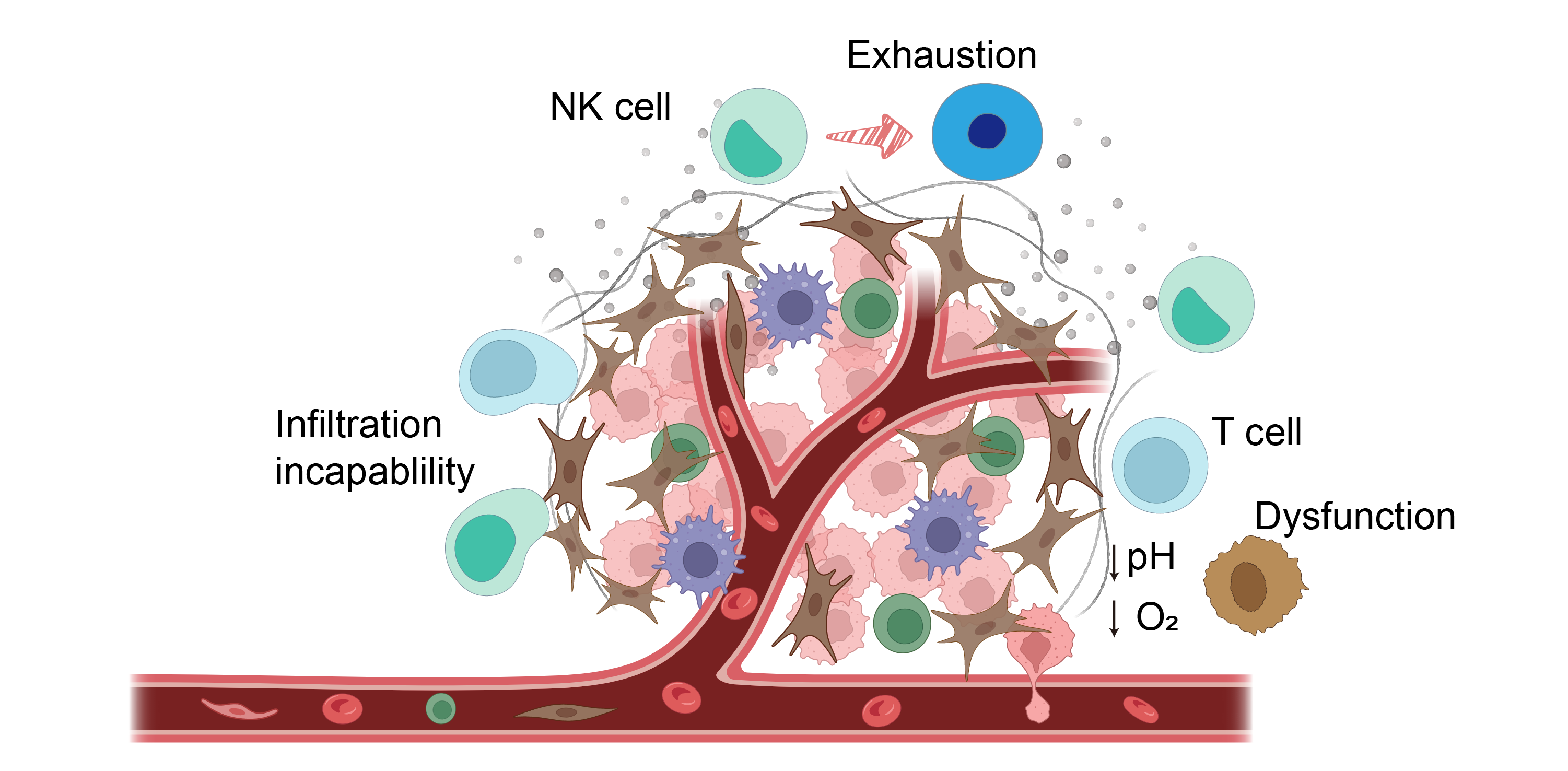
The IL-2 receptor consists of three subunits: IL-2Rα (CD25), IL-2Rβ (CD122), and IL-2Rγ (CD132). Signaling initiation requires a dimeric complex of IL-2Rβ and IL-2Rγ. The incorporation of IL-2Rα into this complex, together with the β and γ chains, forms the high-affinity IL-2R, substantially enhancing IL-2 binding. As a key component of this receptor, CD25 is highly expressed on CD4+CD25+ regulatory T cells (Tregs), enabling their responsiveness to IL-2 and facilitating antigen-nonspecific suppression of effector T cells. Consequently, there is significant interest in developing engineered IL-2 variants that avoid binding to the α-subunit while maintaining affinity for the β and γ subunits. Such variants hold promise for reducing CD25-mediated immunosuppression while preserving the core immunomodulatory functions of IL-2.

Interleukin-2 (IL-2) initiates downstream signaling by binding to the IL-2 receptor (IL-2R), which requires oligomerization of the β chain (IL-2Rβ) and γ chain (IL-2Rγ) to form a stable receptor complex. This oligomerization is critical for the recruitment and activation of intracellular signaling molecules. Upon complex formation, JAK1 and JAK3 are phosphorylated, thereby activating the JAK-STAT pathway, in which phosphorylated STAT5 dimerizes and translocates to the nucleus to regulate transcriptional programs that promote cell proliferation, survival and effector molecule production. Concurrently, the PI3K-AKT pathway is engaged through PI3K recruitment, leading to PIP3 generation and AKT activation, which enhances cell survival, metabolism, and anti-apoptotic signaling.

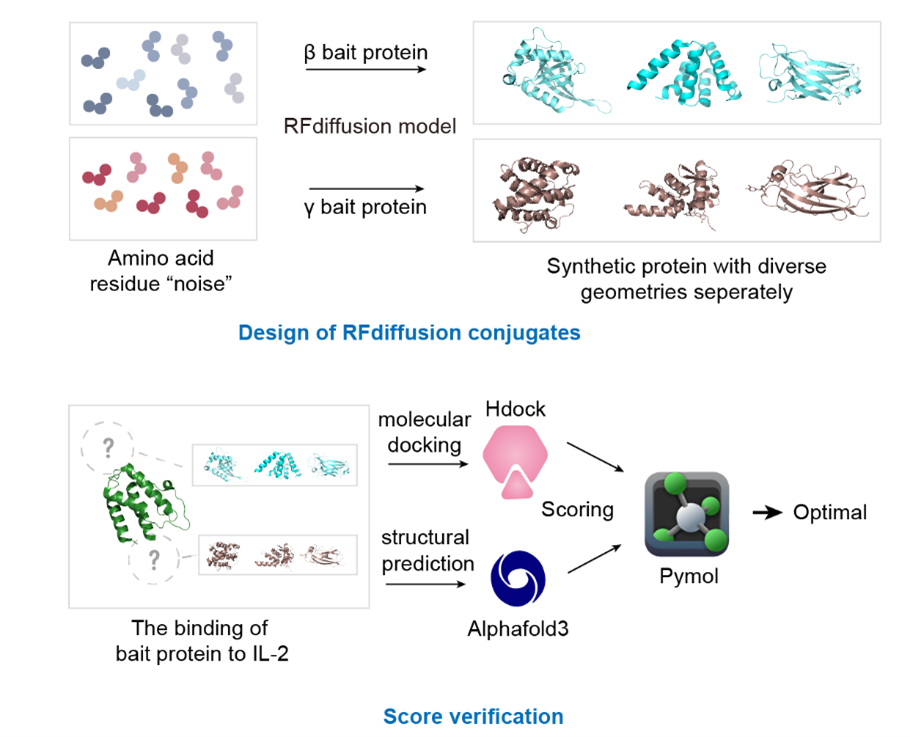
De novo protein design refers to the from-scratch computational generation of protein sequences and structures without relying on naturally occurring templates. It leverages physics-based modeling, energy minimization, and increasingly deep learning algorithms to predict stable folds and desired functions. Representative methods include Rosetta de novo modeling, AlphaFold/AlphaFold3 for structure prediction, and generative models such as ProteinMPNN, RFdiffusion, and EvoDiff. In our project, we designed two distinct libraries of 100 sequences each targeting IL-2Rβ and IL-2Rγ via RFdiffusion. Following the initial HDOCK screening to exclude sequences with high affinity for IL-2Rα, we combinatorially assembled the selected β and γ peptides and then proceeded with structural prediction via AlphaFold3 and experimental validation.

The GALA peptide (WEALLAEALAEALAEHLAEALAEALEALAA) is a rationally designed pH-sensitive sequence. At physiological pH (7.4), the repetitive glutamic acid side-chain carboxyl groups remain negatively charged, and electrostatic repulsion drives the peptide to adopt a random coil conformation. When the pH decreases to 6.4, protonation of the glutamic acid residues reduces electrostatic repulsion, thereby enabling the peptide chain to fold into a rigid α-helical structure. To construct an acid-responsive IL-2R agonist, we incorporated the GALA peptide into the IL-2 mimic.


