1.Wet Lab
1.1 error-prone PCR
The alginate gene we modified was AlyAYO, with a total length of 930bp. We cultured E. coli BL21 containing pET32a-AlyAYO plasmids from its glycerol stock which is from Marine Enzyme Laboratory of Ocean University of China and extracted pET32a-AlyAYO plasmids. Then, we digested pET32a-AlyAYO plasmids with BamHⅠand XhoⅠ to collect pET32a plasmid and AlyAYO gene, which were expression vector and the template of error-prone PCR.
Controlled error-prone PCR Kit (No. ZY-160903) was used to preform sequential error-prone PCR on AlyAYO. The set of number of cycles was 35. The expected mutation values are set to 5 per 1000bp and 8 per 1000bp. In each tube, 5μL of 30μl product was used for agarose gel electrophoresis. The results of error-prone PCR products after agarose gel electrophoresis are shown in Fig1.
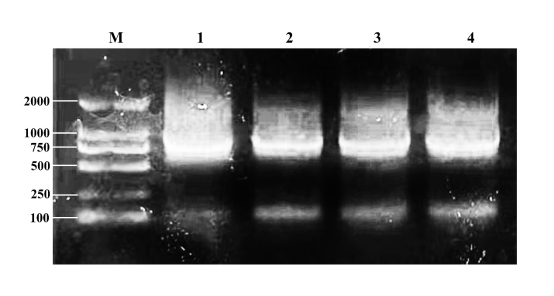
Fig1 Product electrophoresis results
M: Marker2000
1: Error-prone PCR products with a concentration of 10ng/μL as template and the expected mutation value is set to 5 per 1000bp
2: Error-prone PCR products with a concentration of 1ng/μL as template and the expected mutation value is set to 5 per 1000bp
3: Error-prone PCR products with a concentration of 10ng/μL as template and the expected mutation value is set to 8 per 1000bp
4: Error-prone PCR products with a concentration of 1ng/μL as template and the expected mutation value is set to 8 per 1000bp
1.2 Construction of expression vector
The remaining product was digested sequentially with BamHⅠand XhoⅠ. Then we ligated error-prone PCR product into the corresponding sites of the pET32a plasmid to construct the vector and the vector was transformed into E. coli BL21 which was self-made by Marine Enzyme Laboratory of Ocean University of China.
1.3 Enzymatic activity analysis of expressed proteins
We picked single colonies which are successfully transformed and cultured them in 96 well plates. After induction with IPTG, we lysed the cells, used the supernatant as the crude enzyme solution, and measured the enzyme activity.
One unit of enzyme activity was defined as an increase of 1 OD235 unit per min. The enzyme activity of original strain was 12.350U/ml. Among the 32 mutant strain, the enzyme activity of some of them have significantly increased, which are shown as follows. The enzyme activity of the mutant with the highest activity is more than three times that of the wild type.
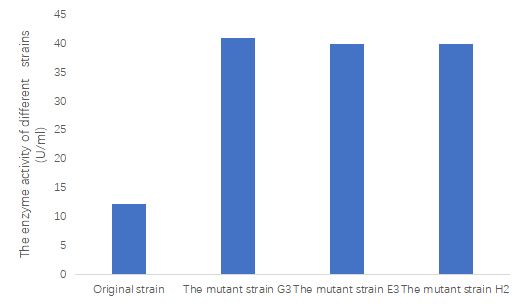
Fig2 The enzyme activity of original strain and its mutant strains.
2. Dry Lab
2.1 A positively mutant protein R30W
Finally, we obtained a positively mutant protein R30Wwhose amino acid sequence is GPPDLGTDDDDKAMADIASDFPNNKETGEALLTPVDATASSHDGNGPDWLIDQDLTTRWSSAGDGEWAMLDYGSVQEFDAVQASFSKGNERQSKFDIQVSVDGETWTTVLENQLSSGKAIGLERFQFEPAVKARYVRYVGHGNTKNGWNSVTGLAAVNCSINACPASQIITSDVVAAEAVLIAEMKAAEKARKAARKDLRSGNFGVAAVYPCETSVKCDTRSALPVPTGLPATPVAGNAPSENFDMTHWYLSQPFDHDKNGKPDDVSEWNLANGYQHPEIFYTADDGGLVFKSYVKGVRTSKNTKYARTELREMMRRGDQSISTKGVNKNNWVFSSAPEADLEAAAGIDGVLEATLKIDHATTTGNANEVGRFIIGQIHDQNDEPIRLYYRKLPNQPTGAVYFAHESQDATKEDFYPLVGDMTAEVGEDGIALGEVFSYRIDVKGNTMTVTLMREGKDDVVQVVDMSNSGYDVGGKYMYFKAGVYNQNISGDLDDYSQATFYQLDVSHDQYKK. Compared with 5ZU5, the docking score of R30W has not changed, but the number of binding sites with sodium alginate has increased from 25 to 37. More binding sites may enhance the interaction between protein and ligand, resulting in enhanced binding affinity, thereby improving the biological activity of the protein. The 3D docking molecule structure of the R30W is shown in Figure 3.
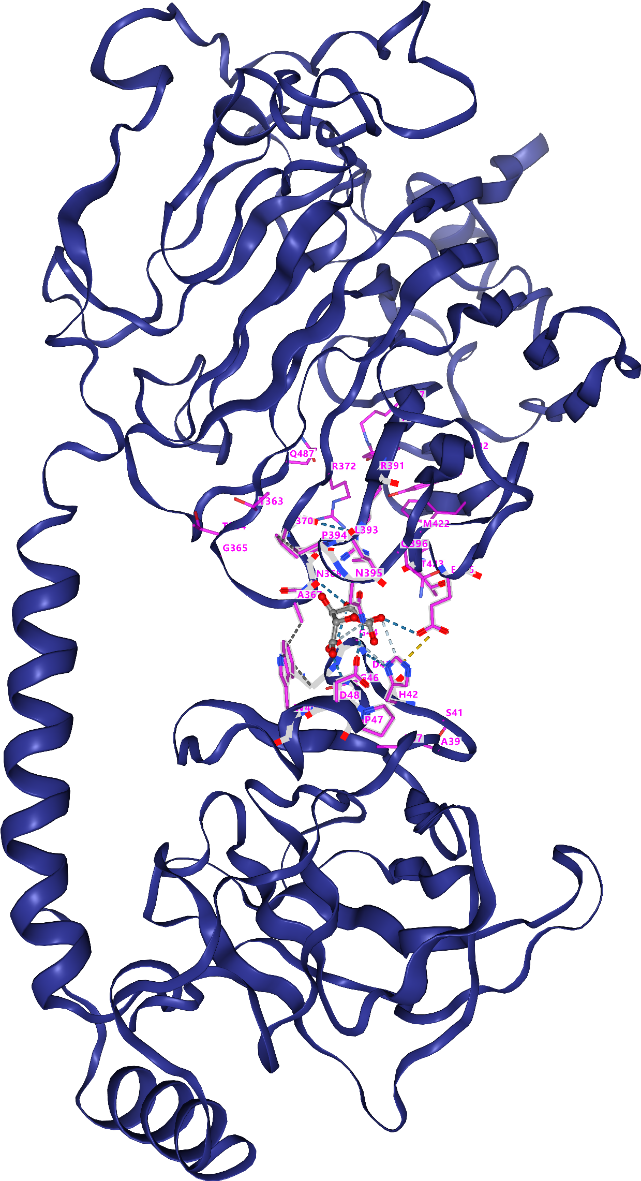
Fig3The 3D docking molecule structure of the R30W.
The amino acids marked in purple in the figure are the amino acids that bind to the substrate, which are ASP36, ALA37, THR38, ALA39, HIS42, ASP43, GLY44, ASN45, GLY46, PRO47, ASP48, TRP49, LYS305, TYR306, THR363, ALA367, ASN368, GLU369, VAL370, ARG372, ILE374, ARG387, TYR389, ARG391, LEU393, PRO394, ASN395, GLN396, TYR402, ALA404, GLU413, PHE415, MET422, THR423, GLU425, TYR485, GLN487.

