Protocols
General information regarding the listed protocols
- The enzymes used in the experiments were obtained from New England Biolabs. Protocols and enzyme-specific reaction conditions were followed as provided by the manufacturer if not mentioned otherwise.
- QIAprep Spin Miniprep Kit (Qiagen) was used for plasmid isolation from bacterial cultures, as per the protocol provided by the manufacturer.
- Chemicals used for the preparation of buffers, solutions, and other reagents were purchased from Sigma-Aldrich.
- All DNA fragments for the experiments were codon optimised for E. coli, synthesised and ordered from Twist Bioscience.
- Oligonucleotide primers were ordered from Microsynth.
Golden Gate Cloning
Golden Gate constructs were assembled using the EcoFlex MoClo kit1 (Addgene kit #1000000080). This kit comes with level 1 and 2 backbones (see publication for more details).
- Add the following to a PCR tube (use BsaI for level 1 and BsmBI for level 2 assemblies):
| Reagent | Amount |
|---|---|
| Backbone DNA | 50 ng |
| Insert DNA | 100 ng |
| 10 × T4 DNA ligase buffer | 1 μl |
| T4 ligase | 0.5 μl |
| BsaI / BsmBI | 0.5 μl |
| ddH2O | fill up to 10 μl |
- Incubate in the thermocycler using the following protocol:
Level 1 / BsaI
| Step | Nr. of Cycles | Temperature (ºC) | Time |
|---|---|---|---|
| Digestion and ligation | 15-30 | 37 | 10 min |
| Annealing | 15-30 | 16 | 5 min |
| Final digestion and T4 ligase inactivation | 1 | 50 | 5 min |
| Enzyme inactivation | 1 | 80 | 5 min |
Level 2 / BsmBI
| Step | Nr. of Cycles | Temperature (ºC) | Time |
|---|---|---|---|
| Initial digestion | 1 | 42 | 1 h |
| Digestion and ligation | 99 | 42 | 5 min |
| Annealing | 99 | 16 | 1 min |
| Final digestion | 1 | 42 | 1h min |
| Enzyme inactivation | 1 | 80 | 15 min |
Note: Try to use smaller vectors containing fewer inserts as reduced protein expression demands may exhibit higher transformation efficiency due to the reduced size and lower metabolic load on the cells.
Gibson Assembly
- PCR of all inserts and backbones to create appropriate overhangs
- DpnI Digest: add 0.5 μL of DpnI to 25 μL PCR reaction, incubation 15 min at 37°C (to remove the template DNA)
- Gel extraction of the correct PCR products: Follow the protocol of the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
- Mix 50–100 ng of vector DNA with a molar 2:1 or 1:1 ratio of each insert (see table below)
- Add water and NEBuilider HiFi DNA Assembly Master Mix as indicated below
| Reagent | 2-3 Fragment Assembly | 4-6 Fragment Assembly |
|---|---|---|
| molar DNA ratio (vector:insert) | 1:2 | 1:1 |
| total amounts of DNA fragments | 0.03-0.2 pmol | 0.2-0.5 pmol |
| NEBuilider HiFi DNA Assembly Master Mix | 2.5 μl | 2.5 μl |
| ddH2O | fill up to 5 μl | fill up to 5 μl |
- Incubate samples in a thermocycler at 50°C for 60 minutes
- Store samples on ice or at –20°C for subsequent transformation
Bacterial transformation
To test our genetically engineered constructs, we transformed them into bacteria. For single plasmid transformation, E. coli cells were made chemically competent using the Mix & Go E. coli Transformation Kit & Buffer Set (Zymo Research), following the manufacturer's protocol.
- Thaw competent DH5a on ice
- Add 1 μl of plasmid DNA or 5 μL of assembly reaction to 20-50 μl of competent cells
- Flick the tube to mix
- Incubate mixture on ice for 30 minutes
- Heat shock at the bacteria at 42°C for 30 s
- Place on ice for 5 minutes
- Pipette 200 µl of room temperature LB into the mixture
- Incubate at 37°C for 60 minutes while shaking at 750 rpm
- Warm selection plates to 37°C in the incubator
- Spin down bacteria and resuspend in 50 μl LB
- Spread 50-100 μl of cells on LB agar plates containing the antibiotics for selection. When multiple antibiotics are used, use 0.6x of the standard working concentration for each antibiotic.
- Incubate overnight at 37°C
For co-transformations with two accessory plasmids: 1. Thaw competent S2060 cells on ice 2. Pre-mix 200 ng of the two accessory plasmids in an equimolar ratio 3. Add 50 μl of competent cells 4. Continue at step 3 above
Due to the metabolic burden caused by DNA replication and protein expression, co-transformants might grow at a markedly reduced rate.
Phage production
The production of our phages started by assembling a plasmid containing all phage genes. Our selection plasmid, containing either SIAH1/2 or unspecific enzymes but lacking essential genes for phage replication was combined with splitC (Addgene #138523) and splitD (Addgene #138521) plasmids using SapI Golden Gate assembly.
Day 1: Golden Gate assembly
Assemble a plasmid containing all phage genes: the selection plasmid, splitC (Addgene #138523) and splitD (Addgene #138521) by mixing the following components in a PCR tube
| Reagent | Amount |
|---|---|
| Selection plasmid | 200 ng |
| SplitC | 100 ng |
| SplitD | 100 ng |
| 10 × T4 ligase buffer | 1 μl |
| T4 ligase | 0.5 μl |
| Sapl1 | 0.5 μl |
| ddH2O | fill up to 20 μl |
Incubate in the thermocycler overnight with the following program:
| Step | Nr. of Cycles | Temperature (ºC) | Time |
|---|---|---|---|
| Digestion and ligation | 99 | 37 | 5 min |
| Annealing | 99 | 18 | 5 min |
| Final digestion | 1 | 37 | 1h min |
| Enzyme inactivation | 1 | 80 | 15 min |
Day 2: Transformation
- Transform the assembly reaction as explained above
- Thaw competent S2060 cells on ice
- Add 2 µl of the ligation reaction into 50 μl of competent S2208 cells
- Flick the tube to mix
- Incubate mixture on ice for 30 minutes
- Heat shock at the bacteria at 42°C for 30 s
- Place on ice for 5 minutes
- Pipette 200 µl of room temperature LB into the mixture
- Incubate at 37°C while shaking at 750 rpm until bacteria reach saturation
- Warm selection plates to 37°C in the incubator
- Spin down bacteria and resuspend in 50 μl LB
- Spread 50-100 μl of cells on LB agar plates
- Incubate overnight at 37°C
Day 3: Plaque Assay to determine phage titer
- Spin down 2 mL (9000 xg, 1 min)
- Transfer supernatant to a new tube and sterilise with a 0.22 µm filter
- Send purified phages for Sanger sequencing with the primer oLS670
- Determine the phage titer with a plaque assay as described below
- Phages can be stored in the fridge for multiple months. If titer gets to low, re-grow in S2208 bacteria again
Note: it is recommended to do 2 plaque assays. The first assay is to determine if the phage production was succesful. From the first assay plate, one plaque is picked to get monoclonal phages and a second assay is performed to assess the PFU.
Determination of phage titer by plaque assay
- Spin down your samples at 8'000 xg for 2 min to remove bacteria and other contaminants
- Transfer supernatant to a new tube. Phages can be stored at 4ºC for further use (note that phage titer will gradually decrease over time)
- Dilute a saturated S2208 culture and grow it to an OD600 of 0.5
- Pour 15 mL LB agar without antibiotics in a Petri dish and let solidify
- Once solidified, mix 2 mL of LB and 2 mL of hot LB agar, add 1 mL of the S2208 culture, vortex quickly, and pour on top of the solidified LB agar. Let solidify for at least 30 min
- Meanwhile, prepare a 10-fold serial dilution of your phage sample (100 to 10-7). Remember to include a phage with a known pfu/mL as a positive control
- Add 4 µL of each dilution onto the top agar. Let the plate sit for 15-30 min before transferring it to the incubator, making sure that the drops do not merge
- Incubate plate overnight (16-20 h) at 37ºC
- The tier of the phage sample can be determined by the following formula:
titer (in pfu/mL) = (# of plaques) x (dilution factor) x 100
Determination of phage titer by qPCR
- Spin down your samples at 8'000 xg for 2 min to remove bacteria and other contaminants
- Transfer supernatant to a new tube. Phages can be stored at 4ºC for further use (note that phage titer will gradually decrease over time)
- Prepare the qPCR reaction mix as follows:
| Reagent | Volume (µL) |
|---|---|
| FirePol Polymerase | 3 |
| 100 uM Forward Primer* | 0.05 |
| 100 uM Reverse Primer* | 0.05 |
| ddH2O | 9.9 |
*oLS-1662 100 uM and oLS-1663 were used as forward and reverse primers respectively, see Supplementary Table 4 for sequences.
- In a qPCR 384-well plate, add 13 µL of qPCR reaction mix to each well, followed by 2 µL of the supernatant of each sample. Perform triplicate measurements for better results
- Cover the plate with an adhesive seal and briefly spin down the plate
- Insert the plate in the qPCR machine (here, we used the LightCycler 480 II from Roche) and program the following settings:
| Step | Nr. of Cycles | Temperature (ºC) | Time |
|---|---|---|---|
| Pre-incubation | 1 | 95 | 12 min |
| Denaturation | 45 | 95 | 15 s |
| Annealing | 45 | 53 | 30 s |
| Elongation | 45 | 72 | 10s |
- Set the excitation wavelength to 465 nm and the detection wavelength to 510 nm and start the protocol
- Analyse readout with the software analysis tool “Absolute Quantification/Second Derivative Maximum”
- Threshold cycle (CT) values, which indicate the cycle number at which the fluorescence from the amplifying DNA exceeded a predefined threshold, can be converted to PFU/mL using a previously constructed standard curved. qPCR results with CT > 30 can be considered as absence of phages
Site-directed mutagenesis for substrate degron modification
- Prepare a reaction mix in a PCR tube as indicated here (see Report for the corresponding primers):
| Reagent | Ammount |
|---|---|
| 5 × Q5 Reaction Buffer | 5 µL |
| 10 µM Forward Primer | 1.25 µL |
| 10 µM Reverse Primer | 1.25 µL |
| pES1076 template | 100 ng |
| Q5 High-Fidelity DNA Polymerase | 0.25 µL |
| Nuclease-Free Water | fill up to 25 µL |
- Gently mix the reaction and collect all liquid to the bottom of the tube by briefly spinning it, if necessary Set the thermocycler for the following program:
| Step | Nr. of Cycles | Temperature (ºC) | Time |
|---|---|---|---|
| Initial Denaturation | 1 | 98 | 30 s |
| Denaturation | 30 | 30 | 5 s |
| Annealing | 30 | 50-72* | 20 s |
| Elongation | 30 | 72 | 20 s/kb of product |
| Final Elongation | 1 | 72 | 2 min |
*calculate annealing temperature for your pair of primers with the NEB Tm Calculator
- Treat 1 µL of the PCR reaction with 5 µL of KLD buffer, 1 µL of the KLD Enzyme Mix, and top it with 3 µL of water
- Incubate at room temperature for 5 min
- Transform 2 µL of the KLD reaction into 20 µL of competent cells and plate directly in LB agar with the appropriate antibiotics without recovery
Phage propagation assay
- Transform S2060 cells with the AP(s) of interest (see Transformation)
- Dilute a saturated overnight culture of bacteria 1,000-fold into LB media supplemented with the appropriate antibiotics and grow at 37ºC to OD600 0.5
- Aliquote 1 mL of culture into a deep-well plate and infect with phages at a starting titer of 106 pfu/mL. Seal the plate with a breathable membrane.
- Incubate the plate overnight (16-20 h) at 37ºC
- Determine the phage titer by performing a plaque assay (see Determination of phage titer by plaque assay). To calculate fold propagation, divide the final phage titer by the starting titer
Phage-assisted non-continuous evolution (PANCE) to generate a library of Selection Plasmids (SP)
- To drift your selection plasmid, you will use a S2060 strain transformed with DP6 plasmid (see Transformation)
- Dilute a saturated overnight bacteria culture in 50 mL LB with appropriate antibiotics and 100 mM glucose, and grow it to an OD600 of 0.5
- Add arabinose to a final concentration of 40 mM. This will induce the expression of the mutagenesis machinery.
- Immediately after, infect your culture with phages containing the SP you want to drift to a final concentration of 104 pfu/mL
- Incubate the flask at 37ºC, shaking at 250 rpm, overnight (16-20 h)
- The following day, determine the phage titer using qPCR (see Determination of phage titer by qPCR)
- Initiate the next round of drift by repeating steps 2 to 6. Infect the fresh culture with the phages that propagated overnight and whose titer you determined by qPCR
Phage-assisted continuous evolution (PACE)
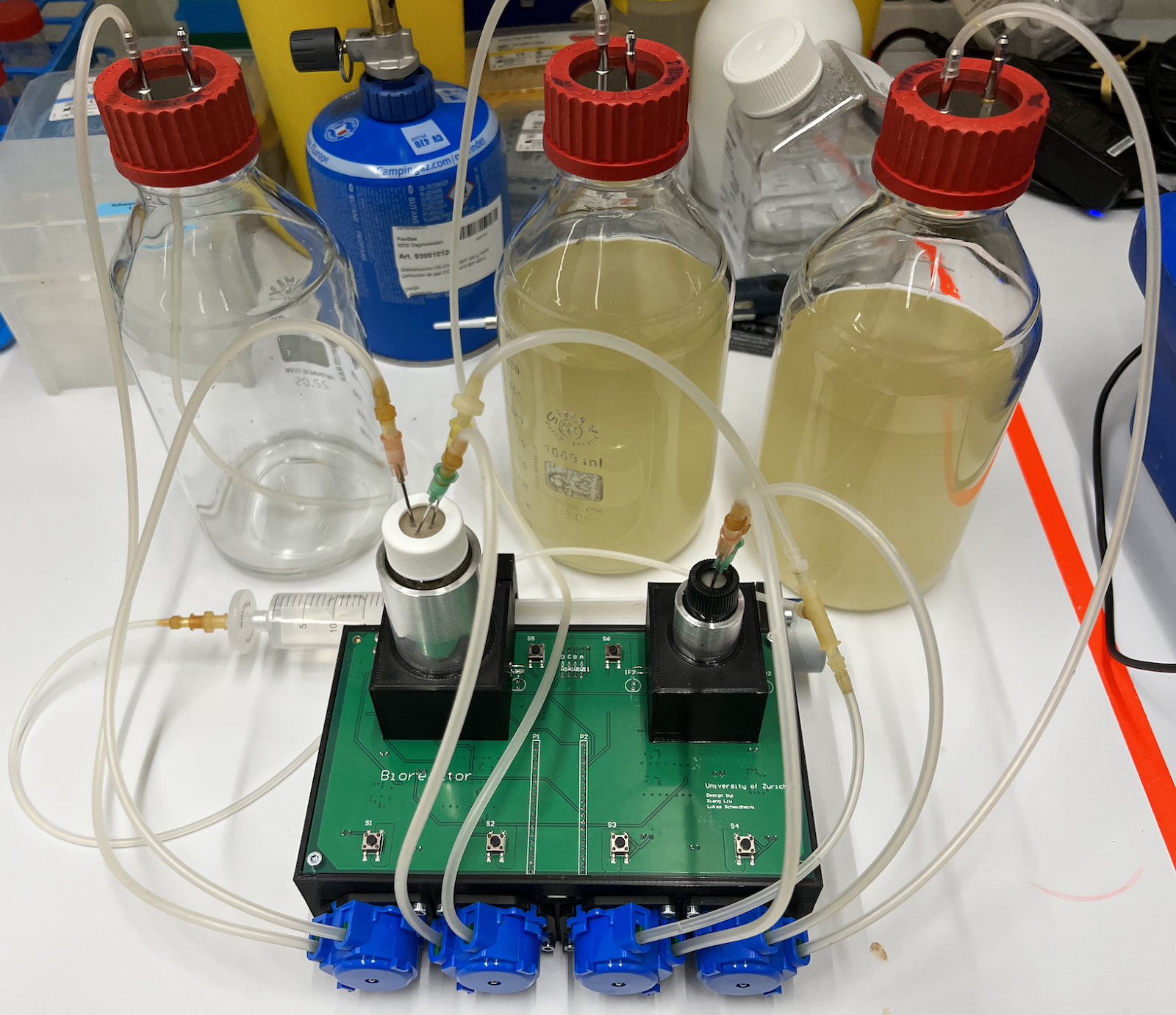
Setting up the PACE reactior
- Autoclave all parts (media bottle, tubing with adaptors, glass bottles with correct stir bar for Chemostat and lagoon). Close any openings with aluminium foil and secure the foil with tape. Autoclave LB with Tube already inserted.
- Inoculate 30 mL LB with bacteria (correct stringency, correct media depending on strain), grow to OD600 of 0.6 (takes about 8 h). Bacteria can be stored in the fridge overnight.
- Important: Work in sterile conditions when assembling the reactor to avoid contaminations. All components should be either autoclaved or sterile already.
- Add antibiotics/inducers etc. to media bottle
- Set up hardware: connect tubing and insert tubes into pump, connect waste and media bottles (see figure). Make sure the orientation of the tubing in the pumps is correct.
- Prepare glass bottle for chemostat (transfer 30 mL of liquid culture grown the day before). Mark the liquid level with a pen on the bottle.
- Insert the three needles in the lagoon
- Waste: insert it at the edge. It should touch the liquid surface and put it against the glass wall
- Fresh media: should be inserted at the centre, higher than the waste needle
- Air: only insert it a bit (highest needle), connect a sterile filter on the outside.
- Run Chemostat until the correct OD is reached. (e.g. overnight)
- Set up lagoon:
- add phages (the titer should be around CT = 25).
- Connect lagoon to chemostat: Tubing to and from lagoon, Lagoon waste
- Insert the three needles in the chemostat and one in the lagoon
- Tubing from chemostat to lagoon: The needle in the chemostat should be inserted all the way (lowest needle) to ensure that there are always bacteria flowing to the reactor whereas the needle in the lagoon should not touch the liquid surface.
- Lagoon to waste: insert it at the edge. It should touch the liquid surface and put it against the glass wall (at 8 mL mark).
- Lagoon to air: only insert it a bit (highest needle), connect a sterile filter on the outside.
- Connect Arabinose: fill syringe with Arabinose, attach a sterile filter to the tip & attach it to the tubing (in front of the pump 3).
- Start PACE. Without arabinose until the lagoon is filled, then start arabinose flow.
Maintaining PACE
The reactor needs to be checked every day to make sure PACE is running smoothly. 1. Check medium, waste and arabinose levels. Replace if necessary. 2. Determine phage titer by qPCR every 12-24 h 3. Check that the reactor is running properly: * Check that stirring bars are stirring and that turbidostat is warm (should be 37°C) * Check medium consumption since the last night * Check amount of disposed lagoon waste from the last night 7. Mark new levels of medium and lagoon waste
Data analysis
General data analysis and visualisation
Raw data and code used to analyse it are available on a public GitHub repository.
Analysis of phage drift using Sanger sequencing
Data were analysed and visualised using R Statistical Software v4.4.12. Sanger sequencing result files were processed using the sangerseqR package v1.40.03. For each nucleotide position, the fraction of mutated signal was calculated as the ratio of the maximum peak amplitude for each non-called base to the total peak amplitudes within the respective base-calling window. To account for signal degradation commonly observed at the extremities of Sanger sequencing reads, only positions between 50 and 650 were considered.
References
-
Moore SJ, Lai H-E, Kelwick RJR, Chee SM, Bell DJ, Polizzi KM, et al. EcoFlex: A multifunctional MoClo kit for E. coli synthetic biology. ACS Synth Biol. 2016;5: 1059–1069. doi:10.1021/acssynbio.6b00031 ↩
-
Komelj J. R Core team: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. In: Scientific Research [Internet]. 2023 [cited 1 Oct 2024]. Available: https://www.scirp.org/reference/referencespapers?referenceid=3582659 ↩
-
Hill JT, Demarest BL, Bisgrove BW, Su Y-C, Smith M, Yost HJ. Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev Dyn. 2014;243: 1632–1636. doi:10.1002/dvdy.24183 ↩