PHB
This sub-section of the project goes over our efforts to optimize PHB production in our cells. See our background section for more context.
PHB Biosynthesis
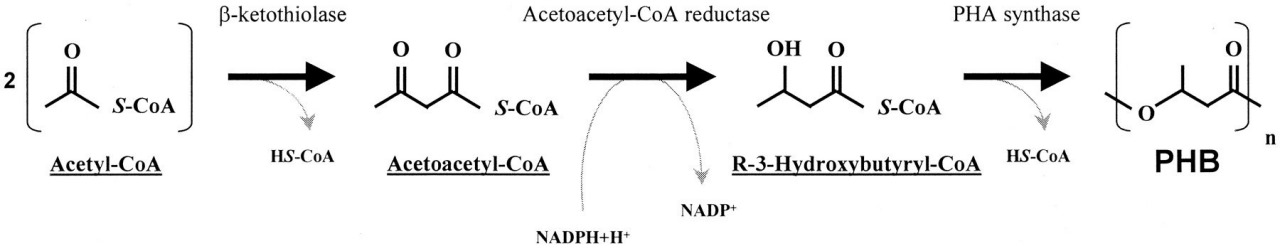
Examining the biosynthesis pathway of Polyhydroxybutyrate (PHB) in Synechocystis sheds light on the pivotal role of acetyl-CoA in a three-step reaction leading to PHB production (Fig 2). The process initiates with 3-ketothiolase catalyzing the Claisen condensation of two acetyl-CoA molecules, yielding acetoacetyl-CoA. Subsequently, acetoacetyl-CoA transforms into R-3-hydroxybutyryl-CoA under the influence of acetoacetyl-CoA reductase. Finally, PHA synthase orchestrates the polymerization of R-3-hydroxybutyryl-CoA. Genes phaA (3-ketothiolase), phaB (acetoacetyl-CoA), phaC, and phaE (PHA synthase subunits) encode the enzymes for these critical steps.
In the pursuit of enhancing PHB production, attention has been directed towards the phaB gene. This avenue was explored by overexpressing the PHA synthase using the Ralstonia eutropha operon in Synechocystis sp. PCC6803. While the PHA synthase witnessed a two-fold increase in expression, the resulting PHB content only exhibited marginal improvement. This observation suggests that the polymerization reaction catalyzed by PHA synthase may not be the primary rate-limiting step in the PHB synthesis pathway.
Overexpressing the native pha operon in Synechocystis PCC6803 led to the highest PHB productivity. Notably, the genes phaA and phaB were located in the same operon, and focusing on the phaB gene contributed to the success of this approach. This strategy, which emphasizes the phaB gene, holds promise for further optimizing PHB production through targeted genetic modifications in the biosynthesis pathway.
Design
Error-prone PCR is a commonly use approach to generate mutant libraries using low-fidelity DNA polymerases such as Taq M0267 lacking 3’-5’ exonuclease proof-reading abilities 1 Increasing MgCl2 concentrations further decreases fidelity, increasing mutation rates.
To generate a mutant library for phaB, the rate-limiting gene involved in PHB biosynthesis, a plasmid construct was designed with the complete phaCAB operon. This construct was derived from the iGEM part BBa_K1149051 which contained a hybrid constitutive and native promotor and the phaCAB operon. This part was optimised for bioplastic production in E.coli MG1655 and for directed evolution, the strong promotor would enable easier quantification of PHB production using Nile red fluorescence. We modified the part to contain BsaI overhangs, which would enable us to digest and ligate the ordered G-block into a level 1 plasmid backbone (DVK_AE) from the Cyanogate parts kit we were provided with thanks to the MacCormick lab. Additionally, we included a double terminator (BBa_B0015) in the G-block design. Two primer pairs, one for replicating the plasmid backbone and phaCA, and a second pair for phaB were designed. Two PCR reactions would have been run, one high-fidelity reaction to replicate a linearised backbone for phaCA, and a separate epPCR reaction with low-fidelity polymerase to generate a library of phaB mutants. The two reaction products would then be digested and ligated together to form a circularised level 1 plasmid for phaCAB* where * denotes a mutagenized gene. This would have been transformed into E.coli and expression of PHB would have been quantified by a fluorescent plate reader-based assay using Nile red. Unfortunately, we planned to synthesize the G-block as a level 1 insert from IDT but our level 1 insert was 4.43kb. IDT only synthesizes G-blocks up to 3kb in size and so we were forced to either redesign or abandon this project. Due to the added complexity of having a dual plasmid system with one containing phaCA and another containing phaB, we decided to focus efforts on salinity resistance. The main issue with dual plasmids was that one would likely be in level 1, and another in level T (containing origins of replication for cyanobacteria as well as E.coli) to prevent plasmid loss from the same origin of replication. Additionally, generating a mutant library with both plasmids by co-transformation would have added extra complexity to the project. Due to time pressures and a desire to focus on one directed evolution project, we didn’t follow through with this project.
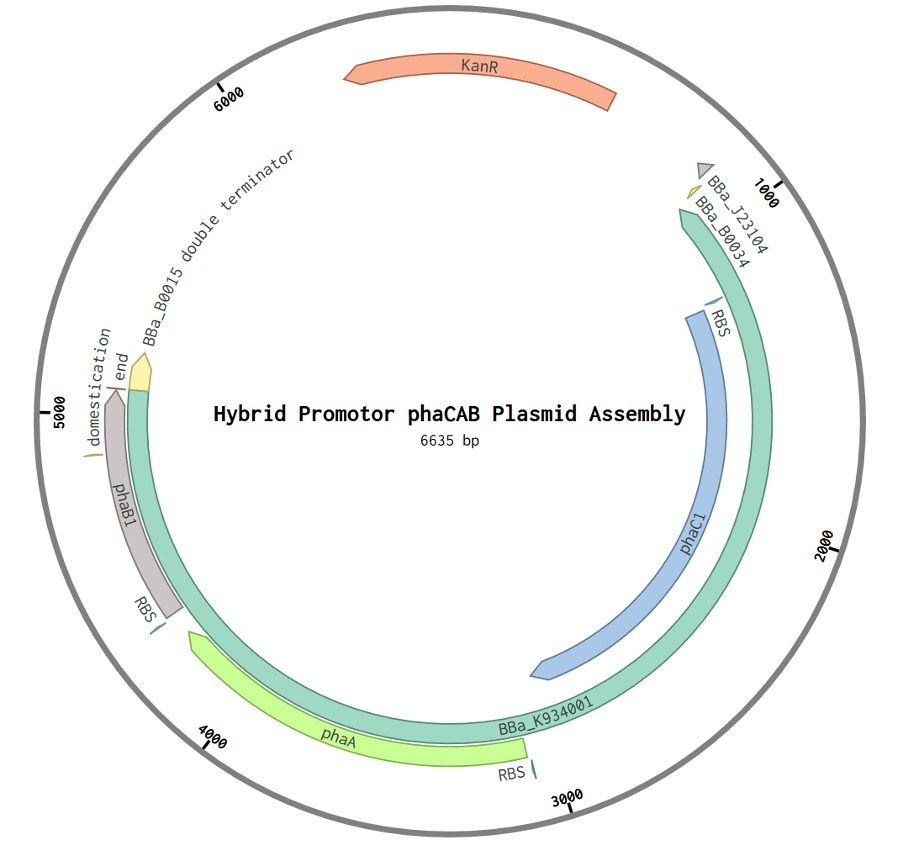
Figure 1: Hybrid Promotor phaCAB Plasmid Assembly. Comprising of the parts BBa_K1149051 and BBa_B0015 inserted into the level 1 plasmid backbone DVK_AE from the Cyanogate kit. A kanamycin selection marker is present (KanR). BBa_J23104 is a constitutive promotor followed by a native promotor. BBa_B0034 is a ribosome binding site.
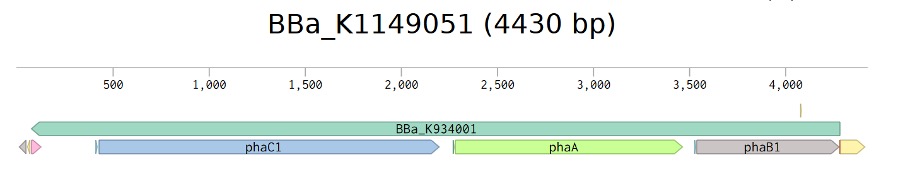
Figure 2: Designed level 1 G-Block for digestion and ligation into level 1 plasmid backbone DVK_AE part colours correspond to figure 1.
| Primer Name | Primer Sequence |
|---|---|
| phaB Forward | cagtGGTCTCaGGACatgactcagcgcattgcgtat |
| phaB Reverse | cagtGGTCTCtCAGGtcagcccatatgcaggccgcc |
| phaCA Backbone Forward | catgGGTCTCtCTGAcctgcccaggcatcaaataa |
| phaCA Backbone Forward | cagtGGTCTCtTCATgtccactccttgattggctt |
Table 1: primer sequences for phaCA and phaB
Results
Nile Red Staining Procedure A volume of 100 μl of wild-type Synechocystic and Synechococcus cells, along with their respective culture media, underwent centrifugation at 1000xg for 1 minute. Subsequently, 80 μl of the supernatant was carefully removed. The pelleted cells were resuspended in a final volume of 20 μl. Following resuspension, the cells were stained with 0.3 μl of Nile Red solution (1.0 μg/ml in ethanol), and an additional 100 μl of 0.9% (w/v) NaCl was introduced. The entire cell population was then incubated in a darkened environment within a cupboard overnight to facilitate optimal staining conditions.
Microscopic Analysis A volume of 10 μl from the stained cyanobacterial media was carefully mounted on a 1% agarose–H2O pad. Brightfield and fluorescence images were captured at a magnification of 0.1 μm per pixel using a Zeiss Axiovert 200 widefield fluorescence microscope, which was outfitted with a Photometrics Evolve 512 EMCCD camera. Subsequent image processing and analysis were performed using Fiji (ImageJ). The selection of the sharpest image plane from the stacks was carried out for a comprehensive representation from a single perspective.
PHB production test with Nile Red The PHB production test for Synechococcus PCC 11901 and Synechocystis PCC6803 involved fluorescent microscopy after Nile red staining, a lipophilic stain commonly used to identify intracellular neutral lipid droplets, including PHB. Both strains were stained to determine if Synechococcus PCC 11901 naturally produces PHB, with Synechocystis PCC6803 as a control. The staining protocol, validated with Synechocystis PCC6803, aimed to screen the native PHB capability in Synechococcus PCC11901. It was expected that PHB would form intracellular granules.
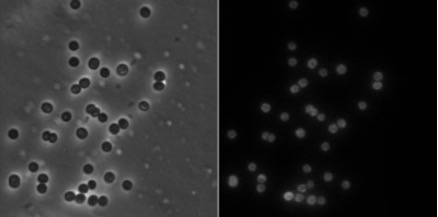
Figure 3: Fluorescent microscopy images of Synechocystis PCC6803 after Nile red staining. Synechocystis PCC6803 were stained by Nile red and visualized by Brightfield(left) and fluorescent microscopy(right).
The fluorescence observed in the cyanobacterial cells under microscopy, aided by ImageJ analysis, revealed a dispersed distribution without identifiable granules. Most cells exhibited reduced fluorescence at the cytoplasm center, possibly due to nucleus presence and PHB storage. The challenge lies in low native cyanobacterial productivity and nonspecific staining distractions. Considering the unknown PHB productivity of Synechococcus PCC11901, optimizing the staining protocol is crucial.
One possible improvement involves inducing nitrogen starvation in cyanobacterial cells, as clear PHB granules may form under such conditions. Experiments with 14-day nitrogen limitation using BG11 and MAD media for Synechocystis PCC6803 and Synechococcus PCC11901, respectively, could provide insights. Staining post-nitrogen starvation and comparing results with cells grown in full-nutrient media may unveil the impact of nitrogen availability on PHB granule visibility. Clear granules in nitrogen-starved cells would suggest its necessity for effective PHB screening. A further improvement may come from adjusting the staining method used to incorporate DSMO which helps permeabilise the lipid membrane. The Nile red dye was likely unable to penetrate the cells sufficiently and so membrane-bound Nile red overshadowed any PHB bound intracellular Nile red.