Lab notebook
Week 1 (19-25 June) Preparation for E.coli K12 growth curves.
Prepared a fully saturated solution of NaCl and did a preliminary E.coli K12 growth curve taking the OD of an inoculated nutrient broth once every half hour for six hours. Prepared a set of nutrient agar plates with a salt gradient ranging from 0.5-5% and plated E.coli K12. E.coli colonies were visible at all concentrations up to 5% which was not expected as literature suggested that they stopped growing at 1.5%. Light microscope analysis indicated that colonies on plates with concentrations higher than 3% NaCl had lysed.
Week 2 (26 Jun -2 July) Designed and ordered primers to extract ggpS and stpA from Synechocystis sp. PCC 6803 genome.
Week 3 (3-9 July) Did PCR to extract ggpS and stpA. ggpS was obtained on the first try, but we had to set a temperature gradient to find the optimum annealing temperature for stpA. Both genes were successfully PCR and gel purified by the end of the week, but the concentrations were not high enough for us to transform into level 0.
Week 4 (10 - 17 July) Repeated PCR extraction of ggpS and stpA to create a more concentrated product, but ran into some problems because the ggpS PCR was no longer working. After repeating the experiment a few times, we managed to get purified ggpS and stpA of high enough concentration.
We performed a level 0 assembly to insert ggpS and stpA separately into pre-mini prepped pICH41308 plasmids. We then transformed these plasmids into TOP10 cells. The ggpS transformation had white colonies, but the stpA transformation was unsuccessful. We picked successful ggpS colonies for colony PCR and the tubes were frozen over the weekend.
Week 5 (18-23 July) We performed gel electrophoresis on frozen colony PCR tubes to see the results of the ggpS level 0 transformation. 5/8 colonies yielded correct band sizes and all 5 colonies were streaked onto plates with IPTG and Xgal. The stpA level 0 assembly and transformation was repeated but unsuccessful.
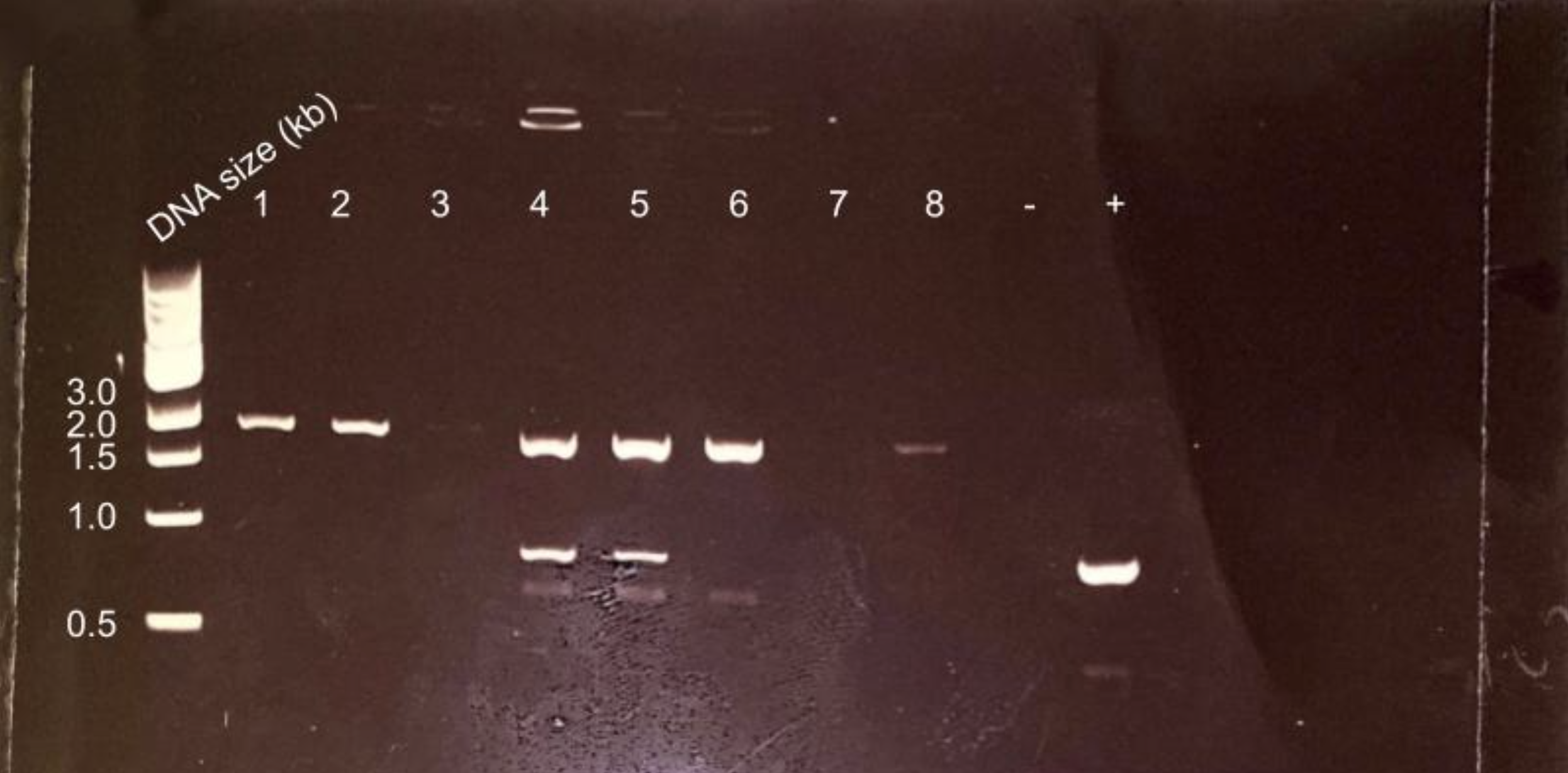 Agarose gel electrophoresis of colony PCR of ggpS level 0 reaction. Correct bands for colonies 1,2,4,5,6,8.
Agarose gel electrophoresis of colony PCR of ggpS level 0 reaction. Correct bands for colonies 1,2,4,5,6,8.
Week 6 (24-30 July) Attempted again the stpA level 0 reaction, increasing the concentration of stpA insert from 2x the concentration of the level 0 vector to 5x the concentration of the level 0 vector, however this was again unsuccessful.
Week 7 (31 July - 6 Aug) Began synechocystis sp. PCC 6803 growth curve using a variety of salt concentrations ranging from 0.25% to 13% in BG11 media measuring approximately once every 24 hours. While this was ongoing the terminator pC0.082 was miniprepped in preparation for moving ggpS into level 1. However it was noticed that the stock of ptrc10 in our lab had a very low concentration after low nanodrop results and confirmation using a cubit, this was overnight cultured in the incorrect antibiotic - spectinomycin instead of kanamycin. Level 1 reaction was performed using low ptrc10 concentration.
Week 8 (7-13 Aug) The level 1 reaction from the week prior was grown on the lab bench over the weekend with poor growth, a promising colony was picked for colony PCR however the level 1 construct was not present. Ptrc10 promoter was overnight cultured and miniprepped to make fresh stock with higher amounts of DNA. Level 1 reaction attempted again, with transformations and colony PCR however this was also unsuccessful.
Week 9 (14-20 Aug) Miniprepped new ggpS level 0 construct and attempted another level 1 reaction for ggpS. Transformations were done, showing promising colonies and these were picked for colony PCR which showed consistent presence of DNA of the expected size for the ggpS level 1 construct and this was then also confirmed by digestion.
Week 10 (21 -27 Aug) Ligated stpA TWIST insert into pSEVA level T acceptor vector and transformed into TOP10 cells. White colonies were visible and plates were kept over the weekend for colony PCR.
Week 11 (28 Aug - 3 Sep) Colony PCR of stpA level T transformation was successful, all 7 white colonies showed the correct bands. We performed a E.coli BL21 growth curve with nutrient broth.
Week 12 (4 - 10 Sep) We attempted to error prone PCR ggpS from the level 1 assembly using newly designed primers, but were unsuccessful despite troubleshooting multiple factors.
Week 13 (11- 17 Sep) Continued to troubleshoot ggpS epPCR reaction with no success.
Week 14 (18-24 Sep) We attempted a q5 PCR of ggpS from purified ggpS, the level 0 assembly and level1 P1 assembly using new primers. Results showed that the primers worked with all templates but the level 1, indicating there was something wrong with the pICH47732 backbone.
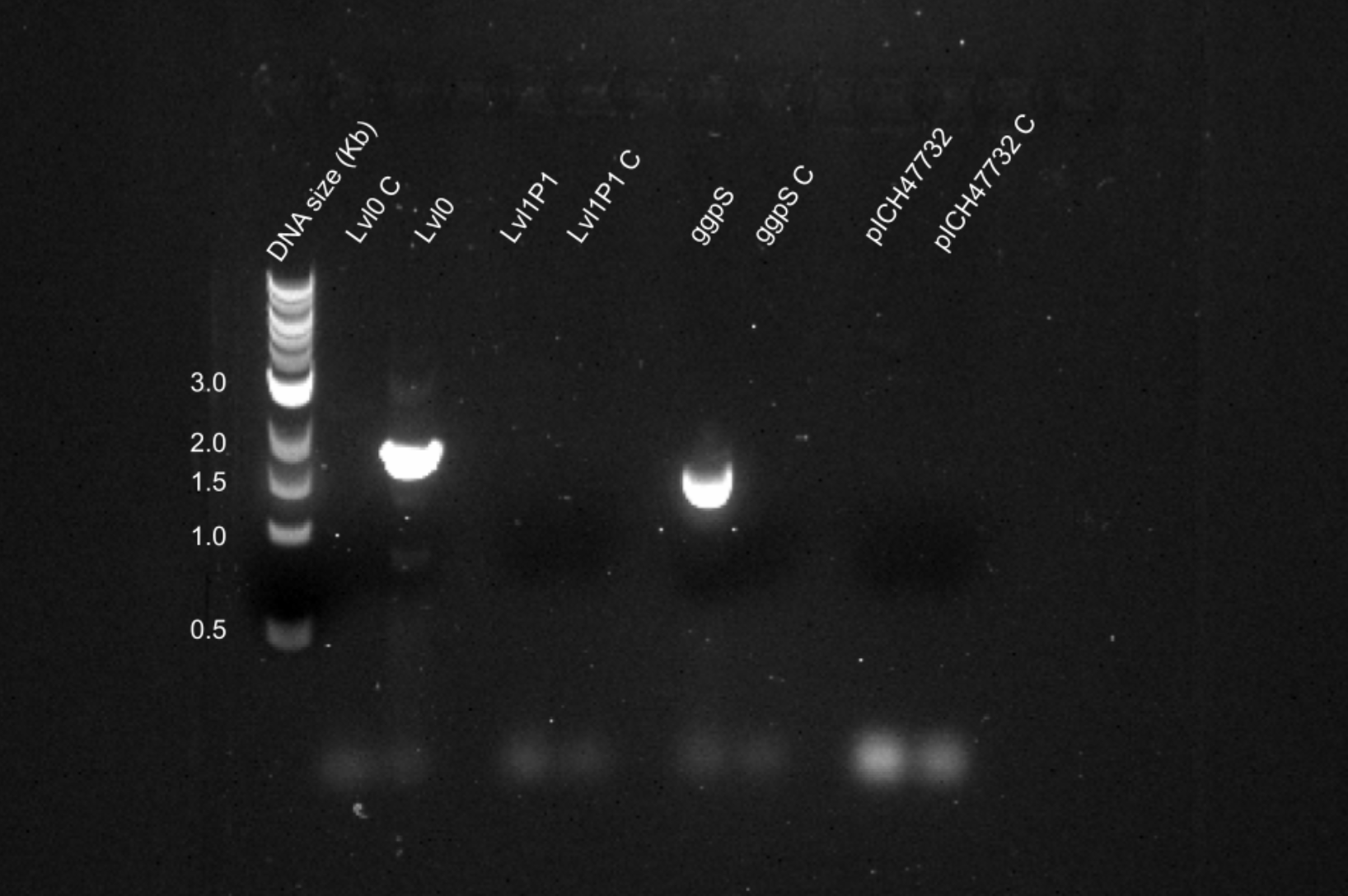 Agarose gel electrophoresis for Q5 PCR of ggpS from ggpS, lvl0 and lvl1P1 templates, and Q5 of pICH47732 backbone from level 1 template.
Agarose gel electrophoresis for Q5 PCR of ggpS from ggpS, lvl0 and lvl1P1 templates, and Q5 of pICH47732 backbone from level 1 template.
Week 15 (25Sep - 1 Oct) We will epPCR the ggpS from the level 0 assembly with the original primers and perform JUMP assembly to insert the library into an e.coli -compatible vector. We will keep some of this library for sequencing to show that the epPCR has worked, and send the JUMP assembly to the edinburgh genome foundry
Week 23 (2-8 Oct)
Transformation of JUMP assembly